Ben-Jacob's bacteria
Ben-Jacob's bacteria are two pattern forming social bacteria strains, the Paenibacillus dendritiformis and the Paenibacillus vortex discovered in the early 1990s by Eshel Ben-Jacob. These bacteria are widely known for their ability to generate large colonies (with the number of bacteria exceeding many folds the number of people on earth) with highly complex organization.
Paenibacillus genus
Ben-Jacob's bacteria belong to the Paenibacillus genus comprises facultative anaerobic, endospore-forming bacteria originally included within the genus Bacillus and then reclassified as a separate genus in 1993.[1] Bacteria belonging to this genus have been detected in a variety of environments such as: soil, water, rhizosphere, vegetable matter, forage and insect larvae, as well as clinical samples.[2][3][4][5] It is mainly found in heterogeneous and complex environments, such as soil and rhizosphere.
In recent years, there is an increasing interest in the Paenibacillus spp. since many were found to be important for industrial, agricultural and medical applications. These bacteria produce various extracellular enzymes such as polysaccharide-degrading enzymes and proteases, which can catalyze a wide variety of synthetic reactions in fields ranging from cosmetics to biofuel production.[6][7][8] Various Paenibacillus spp. also produce antimicrobial substances that affect a wide spectrum of micro-organisms such as fungi, soil bacteria, plant pathogenic bacteria and even important anaerobic pathogens as Clostridium botulinium.[9][10][11]
Physiological and genetic traits
The P. dendritiformis and the P. vortex are facultative anaerobic, lubricating, flagellated and endospore-forming bacteria that are likely to be plant growth promoting rhizobacteria (PGPR). PGPR competitively colonize plant roots and can simultaneously act as biofertilizers and as antagonists (biopesticides) of recognized root pathogens, such as bacteria, fungi and nematodes.[12] They enhance plant growth by several direct and indirect mechanisms. Direct mechanisms include phosphate solubilization, nitrogen fixation, degradation of environmental pollutants and hormone production.
These two pattern forming Paenibacillus strains exhibit many additional distinct physiological and genetic traits including β-galactosidase-like activity causing colonies to turn blue on X-gal plates and multiple drug resistance (MDR) (including septrin, penicillin, kanamycin, chloramphenicol, ampicillin, tetracycline, spectinomycin, streptomycin and mitomycin C. Colonies that are grown on surfaces in Petri dishes exhibit several folds higher drug resistance in comparison to growth in liquid media. This particular resistance is believed to be due to a surfactant-like liquid front that actually forms a particular pattern on the Petri plate.
Social behaviors
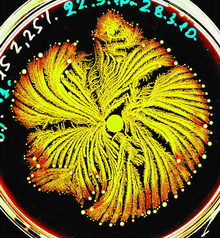
Both P. vortex and P. dendritiformis are social microorganisms: when grown on under growth conditions that mimic natural environments they form colonies with remarkably complex and dynamic architectures (Figures 1 and 2) which behave much like a multi-cellular organism, with cell differentiation and task distribution.[13][14][15][16] P. vortex is marked by its ability to generate special aggregates of dense bacteria that are pushed forward by repulsive chemotactic signals sent from the cells at the back.[17][18][19][20][21][22] These rotating aggregates termed vortices, pave the way for the colony to expand. The vortices serve as building blocks of colonies with special modular organization (Figure 2).
P. dendritiformis, poses an intriguing collective faculty – the ability to switch between different morphotypes[23][24][25] to better adapt with the environment. Mostly studied is the transition between the Branching (or tip-splitting) morphotype and the Chiral morphotype that is marked by curly branches with well defined handedness. The morphotype transitions, can be viewed as an identity switching[23][24][25][26][27] – the calls can cooperatively make drastic alterations of their internal genomic state, effectively transforming themselves into differently looking and behaving cells which can generate colonies with entirely different organization.
Two sibling colonies (colonies taken from the same mother colony or from the same LB growth) of the P. dendritiformis inoculated side by side can inhibit each other from growing into the territory between them and induce the death of those cells close to the border using a special toxin. The latter has been identified and shown to act specifically against the P. dendritiformis.
Communication and chemical tweeting
Accomplishing such intricate cooperative ventures requires sophisticated cell-cell communication[16][23][28][29][30] including semantic and pragmatic aspects of linguistics.[16]
Communicating with each other using a variety of chemical signals, bacteria exchange information regarding population size, a myriad of individual environmental measurements at different locations, their internal states and their phenotypic and epigenetic adjustments. The bacteria collectively sense the environment and execute distributed information processing to glean and assess relevant information. The information is then used by the bacteria for reshaping the colony while redistributing tasks and cell epigenetic differentiations, for collective decision-making and for turning on and off defense and offense mechanisms needed to thrive in competitive environments, faculties that can be perceived as social intelligence of bacteria.[16]
Genome data
The genome sequence of the P. vortex [31] is now available [GenBank: ADHJ00000000. The genome sequence of the P. dendritiformis is now available and will soon be published. Genetic information can be received upon request from the Tauber Sequencing Initiative at Tel-Aviv University.
Analysis of the P. dendritiformis and the P. vortex unveiled the potential to produces a wealth of enzymes and proteases as well as a great variety of antimicrobial substances that affect a wide range of microorganisms. The possession of these advanced defense and offense strategies render these Paenibacillus strains as a rich source of useful genes for agricultural, medical, industrial and biofuel applications.
Bacterial art
The tantalizing beauty of the patterns generated by Ben-Jacob's bacteria made them a widely popular source for bacterial art as the examples shown in Figure 3. These pictures belong to a series of remarkable patterns that Paenibacillus dendritiformis and Paenibacillus vortex bacteria form when grown in a Petri dish under different growth conditions. While the colors and shading are artistic additions, the image templates are actual colonies of tens of billions of these microorganisms. The colony structures form as adaptive responses to laboratory-imposed stresses that mimic hostile environments faced in nature and manipulated by the artist to "guide" the bacteria to generate more complex and artistic patterns.
References
- ↑ Ash C, Priest FG, Collins MD. Molecular identification of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek. 1993;64(3-4):253-260.
- ↑ Lal S, Tabacchioni S: Ecology and biotechnological potential of Paenibacillus polymyxa: a minireview. Indian J Microbiol 2009, 49:2-10.
- ↑ McSpadden Gardener BB: Ecology of Bacillus and Paenibacillus spp. in Agricultural Systems. Phytopathology 2004, 94:1252-1258.
- ↑ Montes MJ, Mercade E, Bozal N, Guinea J: Paenibacillus antarcticus sp. nov., a novel psychrotolerant organism from the Antarctic environment. Int J Syst Evol Microbiol 2004, 54:1521-1526.
- ↑ Ouyang J, Pei Z, Lutwick L, Dalal S, Yang L, Cassai N, Sandhu K, Hanna B, Wieczorek RL, Bluth M, Pincus MR: Case report: Paenibacillus thiaminolyticus: a new cause of human infection, inducing bacteremia in a patient on hemodialysis. Ann Clin Lab Sci 2008, 38:393–400.
- ↑ Konishi J, Maruhashi K: 2-(2'-Hydroxyphenyl)benzene sulfinate desulfinase from the thermophilic desulfurizing bacterium Paenibacillus sp. strain A11-2: purification and characterization. Appl Microbiol Biotechnol 2003, 62:356-361.
- ↑ Raza W, Yang W, Shen QR: Paenibacillus polymyxa: Antibiotics, Hydrolytic Enzymes and Hazard Assessment. J Plant Pathol 2008, 90:419-430.
- ↑ Watanapokasin RY, Boonyakamol A, Sukseree S, Krajarng A, Sophonnithiprasert T, Kanso S, Imai T: Hydrogen production and anaerobic decolorization of wastewater containing Reactive Blue 4 by a bacterial consortium of Salmonella subterranea and Paenibacillus polymyxa. Biodegradation 2009, 20:411-418.
- ↑ Dijksterhuis J, Sanders M, Gorris LG, Smid EJ: Antibiosis plays a role in the context of direct interaction during antagonism of Paenibacillus polymyxa towards Fusarium oxysporum. J Appl Microbiol 1999, 86:13-21.
- ↑ Girardin H, Albagnac C, Dargaignaratz C, Nguyen-The C, Carlin F: Antimicrobial activity of foodborne Paenibacillus and Bacillus spp. against Clostridium botulinum. J Food Prot 2002, 65:806-813.
- ↑ von der Weid I, Alviano DS, Santos AL, Soares RM, Alviano CS, Seldin L: Antimicrobial activity of Paenibacillus peoriae strain NRRL BD-62 against a broad spectrum of phytopathogenic bacteria and fungi. J Appl Microbiol 2003, 95:1143-1151.
- ↑ Bloemberg GV, Lugtenberg BJ: Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr Opin Plant Biol 2001, 4:343-350.
- ↑ Aguilar C, Vlamakis H, Losick R, Kolter R: Thinking about Bacillus subtilis as a multicellular organism. Curr Opin Microbiol 2007, 10:638-643.
- ↑ Dunny GM, Brickman TJ, Dworkin M: Multicellular behavior in bacteria: communication, cooperation, competition and cheating. Bioessays 2008, 30:296-298.
- ↑ Shapiro JA, Dworkin M: Bacteria as multicellular organisms. 1st edn: Oxford University Press, USA; 1997.
- 1 2 3 4 Ben-Jacob E, Becker I, Shapira Y, Levine H: Bacterial linguistic communication and social intelligence. Trends Microbiol 2004, 12:366-372.
- ↑ Ben-Jacob E, Cohen I, Gutnick DL: Cooperative organization of bacterial colonies: from genotype to morphotype. Annu Rev Microbiol 1998, 52:779-806.
- ↑ Ben-Jacob E: From snowflake formation to growth of bacterial colonies II: Cooperative formation of complex colonial patterns. Contem Phys 1997, 38:205 - 241.
- ↑ Ben-Jacob E, Cohen I: Cooperative formation of bacterial patterns. In Bacteria as Multicellular Organisms Edited by Shapiro JA, Dworkin M. New York: Oxford University Press; 1997: 394-416
- ↑ Ben-Jacob E, Cohen I, Czirók A, Vicsek T, Gutnick DL: Chemomodulation of cellular movement, collective formation of vortices by swarming bacteria, and colonial development. Physica A 1997, 238:181-197.
- ↑ Cohen I, Czirok A, Ben-Jacob E: Chemotactic-based adaptive self-organization during colonial development. Physica A 1996, 233:678-698.
- ↑ Czirok A, Ben-Jacob E, Cohen II, Vicsek T: Formation of complex bacterial colonies via self-generated vortices. Phys Rev E Stat Phys Plasmas Fluids Relat Interdiscip Topics 1996, 54:1791-1801.
- 1 2 3 Ben-Jacob E. Bacterial self-organization: co-enhancement of complexification and adaptability in a dynamic environment. Phil. Trans. R. Soc. Lond. A. 2003;361(1807):1283-1312.
- 1 2 Ben-Jacob E, Cohen I, Gutnick DL. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu Rev Microbiol. 1998;52:779-806.
- 1 2 Ben-Jacob E, Cohen I. Cooperative formation of bacterial patterns. In: Shapiro JA, Dworkin M, eds. Bacteria as Multicellular Organisms New York: Oxford University Press; 1997:394-416.
- ↑ Ben-Jacob E, Levine H. Self-engineering capabilities of bacteria. J R Soc Interface. 2005;3(6):197-214.
- ↑ Ben-Jacob E, Cohen I, Golding I, et al. Bacterial cooperative organization under antibiotic stress. Physica A. 2000;282(1-2):247–282.
- ↑ Bassler BL, Losick R: Bacterially speaking. Cell 2006, 125:237-246.
- ↑ Bischofs IB, Hug JA, Liu AW, Wolf DM, Arkin AP. Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proc Natl Acad Sci U S A. Apr 21 2009;106(16):6459-6464.
- ↑ Kolter R, Greenberg EP: Microbial sciences: the superficial life of microbes. Nature 2006, 441:300-302.
- ↑ Sirota-Madi A, Olender T, Helman Y, Ingham C, Brainis I, Roth D, Hagi E, Brodsky L, Leshkowitz D, Galatenko V, et al: Genome sequence of the pattern forming Paenibacillus vortex bacterium reveals potential for thriving in complex environments. BMC Genomics, 11:710.
External links
- Eshel Ben-Jacob's Home Page
- Microbial Art
- Ben Jacob's Fractal Bacteria
- Realizing Social Intelligence of Bacteria
- Bacterial Art
- Bacterial self–organization: co–enhancement of complexification and adaptability in a dynamic environment
- Bacterial linguistic communication and social intelligence
- The genius of bacteria
- Gambling on Bacteria
- The tree of Life: IQ Test for Bacteria