Coronaviridae
| Coronaviridae | |
|---|---|
| Virus classification | |
| Group: | Group IV ((+)ssRNA) |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Subfamilies and Genera | |
Coronaviridae is a family of enveloped, positive-stranded RNA viruses. The viral genome is 26–32 kb in length. Virions are spherical, 120–160 nm across (Coronavirinae), bacilliform, 170–200 by 75–88 nm (Bafinivirus) or found as a mixture of both, with bacilliform particles characteristically bent into crescents (Torovirus). The particles are typically decorated with large (~20 nm), club- or petal-shaped surface projections (the “peplomers” or “spikes”), which in electron micrographs of spherical particles create an image reminiscent of the solar corona. Members of this family are thus referred to as coronaviruses, as are members of the subfamily Coronavirinae.
Virology
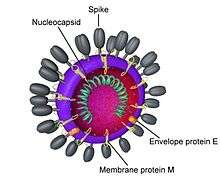
The 5' and 3' ends of the genome have a cap and poly (A) tract, respectively. The viral envelope, obtained by budding through membranes of the ER and/or Golgi apparatus, invariably contains two virus-specified (glyco)protein species, S and M. Glycoprotein S comprises the large surface projections, while the M protein is a triple-spanning transmembrane protein. Toroviruses and a select subset of coronaviruses (in particular the members of subgroup A in the genus Betacoronavirus) possess, in addition to the peplomers composed of S, a second type of surface projections composed of the hemagglutinin-esterase protein. Another important structural protein is the phosphoprotein N, which is responsible for the helical symmetry of the nucleocapsid that encloses the genomic RNA.
Taxonomy
Group: ssRNA(+)
- Family: Coronaviridae
- Sub-Family: Coronavirinae
- Genus: Betacoronavirus
- Betacoronavirus 1
- Human coronavirus HKU1
- Murine coronavirus
- Pipistrellus bat coronavirus HKU5
- Rousettus bat coronavirus HKU9
- Severe acute respiratory syndrome-related coronavirus
- Tylonycteris bat coronavirus HKU4
- Genus: Deltacoronavirus
- Bulbul coronavirus HKU11
- Munia coronavirus HKU13
- Thrush coronavirus HKU12
- Genus: Gammacoronavirus
- Avian coronavirus
- Beluga whale coronavirus SW1
- Sub-Family: Torovirinae
- Genus: Bafinivirus
- Genus: Torovirus
- Bovine torovirus
- Equine torovirus
- Human torovirus
- Porcine torovirus
There are currently a total of 25 species assigned to this family, twenty of which belong to the subfamily Coronavirinae.
Transmission
Coronaviruses are transmitted by faecal-oral route or by aerosols of respiratory secretions.
Pathogenesis
Coronaviruses infect a wide range of mammals and birds and occur worldwide. Although most diseases are mild, sometimes they can cause more severe situations in humans, such as, for example, the infection of the respiratory tract known as Severe Acute Respiratory Syndrome (SARS). They can also cause enteric infections in very young infants and, in rare situations, neurological syndromes.
SARS
Human infection by SARS coronavirus appears to be limited to the respiratory tract where infection of susceptible cells leads to damage to the pneumocytes resulting in a histological picture of diffuse alveolar damage and a clinical picture of adult respiratory distress syndrome. Diarrhea is also present but there is limited evidence of damage to the intestinal epithelium. The damage to the respiratory tree appears limited to the lower respiratory tract and there is evidence that the immune response plays a part in the outcome of patients with SARS.[4]
Binding and entry
Coronaviruses bind to host cells primarily through interactions between viral spike glycoproteins and specific host cell surface glycoproteins. Some coronaviruses also bind to sialic acids on glycoproteins and glycolipids via their spike and/or hemaglutinin esterase glycoproteins. The interactions between coronaviruses and host cell receptors are critical determinants of species-specificity, tissue tropism, and virulence.[2][4]
Replication
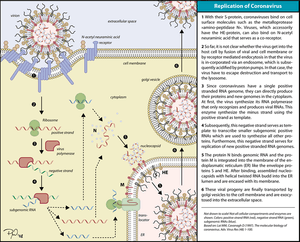
Coronaviruses have single-stranded, positive-sense RNA genomes of 26-30 kilobases, by far the largest non-segmented RNA virus genomes currently known. The key functions required for coronavirus RNA synthesis are encoded by the viral replicase gene. The gene comprises more than 20,000 nucleotides and encodes two replicase polyproteins, pp1a and pp1ab, that are proteolytically processed by viral proteases. Over the past years, it has become clear that the unique size of the coronavirus genome and the special mechanism that coronaviruses (and several other nidoviruses) have evolved to produce an extensive set of subgenome-length RNAs is linked to the production of a number of nonstructural proteins (nsps) that is unprecedented among RNA viruses. Many of these replicase cleavage products are in fact multidomain proteins themselves, thus further increasing the complexity of protein functions and interactions. Structural studies suggest that several nsps, following their release from larger precursor molecules, form dimers or even multimers. The various pp1a/pp1ab precursors and processing products are thought to assemble into large, membrane-associated complexes that, in a temporally coordinated manner, catalyze the reactions involved in RNA replication and transcription and, it is presumed, serve yet other functions in the viral life cycle.[2][4][5]
Coronaviruses also exhibit ribosomal frameshifting and polymerase stuttering as part of their complex replicative cycle.[6][7][8][9][10]
Genomic cis-acting elements
In common with the genomes of all other RNA viruses, coronavirus genomes contain cis-acting RNA elements that ensure the specific replication of viral RNA by a virally encoded RNA-dependent RNA polymerase. The embedded cis-acting elements devoted to coronavirus replication constitute a small fraction of the total genome, but this is, it is presumed, a reflection of the fact that coronaviruses have the largest genomes of all RNA viruses. The boundaries of cis-acting elements essential to replication are fairly well-defined, and an increasingly well-resolved picture of the RNA secondary structures of these regions is emerging. However, we are in only the early stages of understanding how these cis-acting structures and sequences interact with the viral replicase and host cell components, and much remains to be done before we understand the precise mechanistic roles of such elements in RNA synthesis.[2][4]
Genome packaging
The assembly of infectious coronavirus particles requires the selection of viral genomic RNA from a cellular pool that contains an abundant excess of non-viral and viral RNAs. Among the seven to ten specific viral mRNAs synthesized in virus-infected cells, only the full-length genomic RNA is packaged efficiently into coronavirus particles. Studies have revealed cis-acting elements and trans-acting viral factors involved in coronavirus genome encapsidation and packaging. Understanding the molecular mechanisms of genome selection and packaging is critical for developing antiviral strategies and viral expression vectors based on the coronavirus genome.[2][4]
References
- ↑ ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- 1 2 3 4 5 de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJ, Talbot PJ, Woo PC, Ziebuhr J (2011). "Family Coronaviridae". In: Ninth Report of the International Committee on Taxonomy of Viruses. AMQ King, E Lefkowitz, MJ Adams, and EB Carstens (Eds), Elsevier, Oxford, pp. 806-828. ISBN 978-0-12-384684-6.
- ↑ INTERNATIONAL COMMITTEE ON TAXONOMY OF VIRUSES (20 March 2010). "ICTV 2009 MASTER SPECIES LIST VERSION 4"
- 1 2 3 4 5 Thiel V (editor). (2007). Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-16-5. .
- ↑ Enjuanes L; et al. (2008). "Coronavirus Replication and Interaction with Host". Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6.
- ↑ Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. "A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting." Nature. 2006 May 11;441(7090):244-7.
- ↑ Plant, EP; Dinman, JD (Apr 2006). "Comparative study of the effects of heptameric slippery site composition on -1 frameshifting among different eukaryotic systems". RNA. 12 (4): 666–73. doi:10.1261/rna.2225206.
- ↑ Su MC, Chang CT, Chu CH, Tsai CH, Chang KY (2005). "An atypical RNA pseudoknot stimulator and an upstream attenuation signal for -1 ribosomal frameshifting of SARS coronavirus". Nucleic Acids Research. 33 (13): 4265–75. doi:10.1093/nar/gki731. PMC 1182165
 . PMID 16055920.
. PMID 16055920. - ↑ Herrewgh, A.A.; Vennema, H.; Horzinek, M.C.; Rottier, P.J; de Groot, R.J. (1995). "The molecular genetics of feline coronaviruses: comparative sequence analysis of the ORF7a/7b transcription unit of different biotypes" (PDF). Virology. 212 (2): 622–31. doi:10.1006/viro.1995.1520. PMID 7571432.
- ↑ Jeong, YS; Makino, S (Apr 1994). "Evidence for coronavirus discontinuous transcription". J Virol. 68 (4): 2615–23.
External links
- Coronaviridae
- 19. Coronaviridae
- Coronaviruses
- Focus on Coronaviruses
- Viralzone: Coronaviridae
- Virus Pathogen Database and Analysis Resource (ViPR): Coronaviridae
- ICTV