DEPDC5
| DEPDC5 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||||||||
| Aliases | DEPDC5, DEP.5, FFEVF, DEP domain containing 5 | ||||||||||||||||
| External IDs | MGI: 2141101 HomoloGene: 34718 GeneCards: DEPDC5 | ||||||||||||||||
| Genetically Related Diseases | |||||||||||||||||
| hepatitis C[1] | |||||||||||||||||
| |||||||||||||||||
| Orthologs | |||||||||||||||||
| Species | Human | Mouse | |||||||||||||||
| Entrez | |||||||||||||||||
| Ensembl | |||||||||||||||||
| UniProt | |||||||||||||||||
| RefSeq (mRNA) | |||||||||||||||||
| RefSeq (protein) |
| ||||||||||||||||
| Location (UCSC) | Chr 22: 31.75 – 31.91 Mb | Chr 5: 32.86 – 32.99 Mb | |||||||||||||||
| PubMed search | [2] | [3] | |||||||||||||||
| Wikidata | |||||||||||||||||
| View/Edit Human | View/Edit Mouse |
DEPDC5 (or DEP domain-containing 5) is a human protein of poorly understood function but has been associated with cancer in several studies.[4][5] It is encoded by a gene of the same name, located on chromosome 22.
Function
The function of DEPDC5 is not yet known, but it has been implicated in intracellular signal transduction based on homology between the DEP domains of DEPDC5 and Dishevelled-1 (DVL1).[6]
Mutations in this gene have been associated to cases of focal epilepsy (doi:10.1038/ng.2601).
Gene
In Homo sapiens, the DEPDC5 gene has been localized to the long arm of chromosome 22, 22q12.2-q12.3, between the PRRL14 and YWHAH genes. The clinical relevance of this gene includes an intronic SNP (rs1012068) that has been associated with a 2-fold hepatocellular carcinoma-risk increase.[4]

Structure
Domains

DEP
The DEP domain derives its name from the proteins Dishevelled, Egl-10 and Pleckstrin, each of which contain a variant of this domain.[7] It spans 82 residues and is 343 amino acids from the C-terminus. A SWISS-MODEL predicts two beta sheets and three alpha helices contained within the domain.[8]
While its exact function is not known, the DEPDC5 DEP domain has the highest structural similarity to the DEP domain of DVL1 when performing a CBLAST at NCBI.[9] The alignment scores an Evalue of 1.00e-08 and indicates 30% identity between the DEP domains of the two proteins. In DVL1, the DEP domain is involved in localization of the protein to the plasma membrane as part of the Wnt signaling pathway.[10]
DUF 3608
The DUF 3608 domain sits 99 amino acids from the N-terminus and itself spans 280 amino acids. PELE predicts at least one beta sheet and two alpha helices within this domain.[11] It also contains 26 highly conserved residues and several post-translation modifications. Both occurrences are addressed later in this article.
Evidence for the function of DUF 3608 has been uncovered in the yeast homolog Iml1p. Imlp1's DUF 3608 is thought to aid in binding to two protein partners, Npr2 and Npr3. Together, these three proteins form the Iml1-Npr2-Npr3 complex and are involved in "non-nitrogen starvation" autophagy regulation. The researchers who uncovered this propose renaming DUF 3608 to RANS (Required for Autophagy induced under Non-nitrogen Starvation conditions).[12]
Secondary Structure
Based on unanimous consensus by the secondary structure prediction tool PELE, DEPDC5 contains at least ten alpha helices and nine beta sheets. The locations of these secondary structures are illustrated in the image below: red highlights are alpha helices and blue highlights are beta sheets.

Homology
Orthologs
Fungi are the most distantly related organisms to contain a protein orthologous to human DEPDC5, including Saccharomyces cerevisiae and Albugo laibachii. In the fungi, the protein name is Iml1p, or vacuolar membrane-associated protein Iml1. Name deviations in other organisms include CG12090 (Drosophila) and AGAP007010 (mosquito).[6] Conservation is high between humans and other vertebrate species, ranging from 74% identity in cichlids to 99% identity in chimpanzees.[13]
The following table summarizes an analysis of 20 proteins orthologous to human DEPDC5.
| Species | Common Name | NCBI Accession # | NCBI Name | Length | Sequence Identity | Sequence Similarity | Years Since Divergence from Human (mya)[14] |
| Pan troglodytes | Chimpanzee | XP_003317262 | DEPDC5 | 1572 aa | 99% | 99% | 6.4 |
| Nomascus leucogenys | Gibbon | XP_003258163 | DEPDC5 | 1602 aa | 99% | 99% | 20.4 |
| Mus musculus | Mouse | NP_001164038 | DEPDC5 | 1591 aa | 94% | 96% | 92.4 |
| Bos Taurus | Cow | XP_002694678 | DEPDC5 | 1593 aa | 94% | 96% | 94.4 |
| Sorex araneus | Shrew | ACE77702 | DEPDC5 | 1570 aa | 94% | 96% | 94.4 |
| Monodelphis domestica | Possum | XP_001378772 | DEPDC5 | 1522 aa | 89% | 93% | 163.9 |
| Gallus gallus | Chicken | XP_415249 | DEPDC5 | 1592 aa | 88% | 93% | 301.7 |
| Meleagris gallopavo | Turkey | XP_003211073 | DEPDC5 | 1592 aa | 88% | 93% | 301.7 |
| Taeniopygia guttata | Zebra finch | XP_002199825 | DEPDC5 | 1572 aa | 87% | 92% | 301.7 |
| Xenopus tropicalis | Frog | XP_002931964 | DEPDC5-like | 1574 aa | 79% | 86% | 371.2 |
| Danio rerio | Zebra fish | XP_691450 | DEPDC5-like | 1590 aa | 75% | 84% | 400.1 |
| Oreochromis niloticus | Cichlid | XP_003459226 | DEPDC5 | 1577 aa | 74% | 82% | 400.1 |
| Strongylocentrotus purpuratus | Sea urchin | XP_794020 | similar to DEPDC5 | 1608 aa | 43% | 57% | 742.9 |
| Drosophila melanogaster | Drosophila | NP_647618 | GC12090 | 1471 aa | 41% | 57% | 782.7 |
| Pediculus humanus corporis | Louse | XP_002429401 | DEPDC, putative | 1538 aa | 38% | 53% | 782.7 |
| Anopheles gambiae | Mosquito | XP_308760 | AGAP007010-PA | 1640 aa | 36% | 51% | 782.7 |
| Ascaris suum | Ascaris | ADY40551 | DEPDCp5 | 1359 aa | 31% | 51% | 937.5 |
| Ustilago maydis | Corn smut | XP_757759 | vacuolar-associated protein Iml1 | 1867 aa | 23% | 52% | 1215.8 |
| Saccharomyces cerevisiae | Yeast | NP_012672 | Iml1p | 1584 aa | 20% | 50% | 1215.8 |
| Albugo laibachii | White rust | CCA27519 | vacuolar membrane-associated protein putative | 1591 aa | 20% | 46% | 1362 |
30 residues have been conserved since animals and fungi diverged, with 26 of these located in the DUF 3608 domain.[15] The following multiple sequence alignment illustrates this conservation of the DUF domain; representatives from invertebrate and fungal clades are aligned to the human DUF 3608 with completely conserved residues colored green.
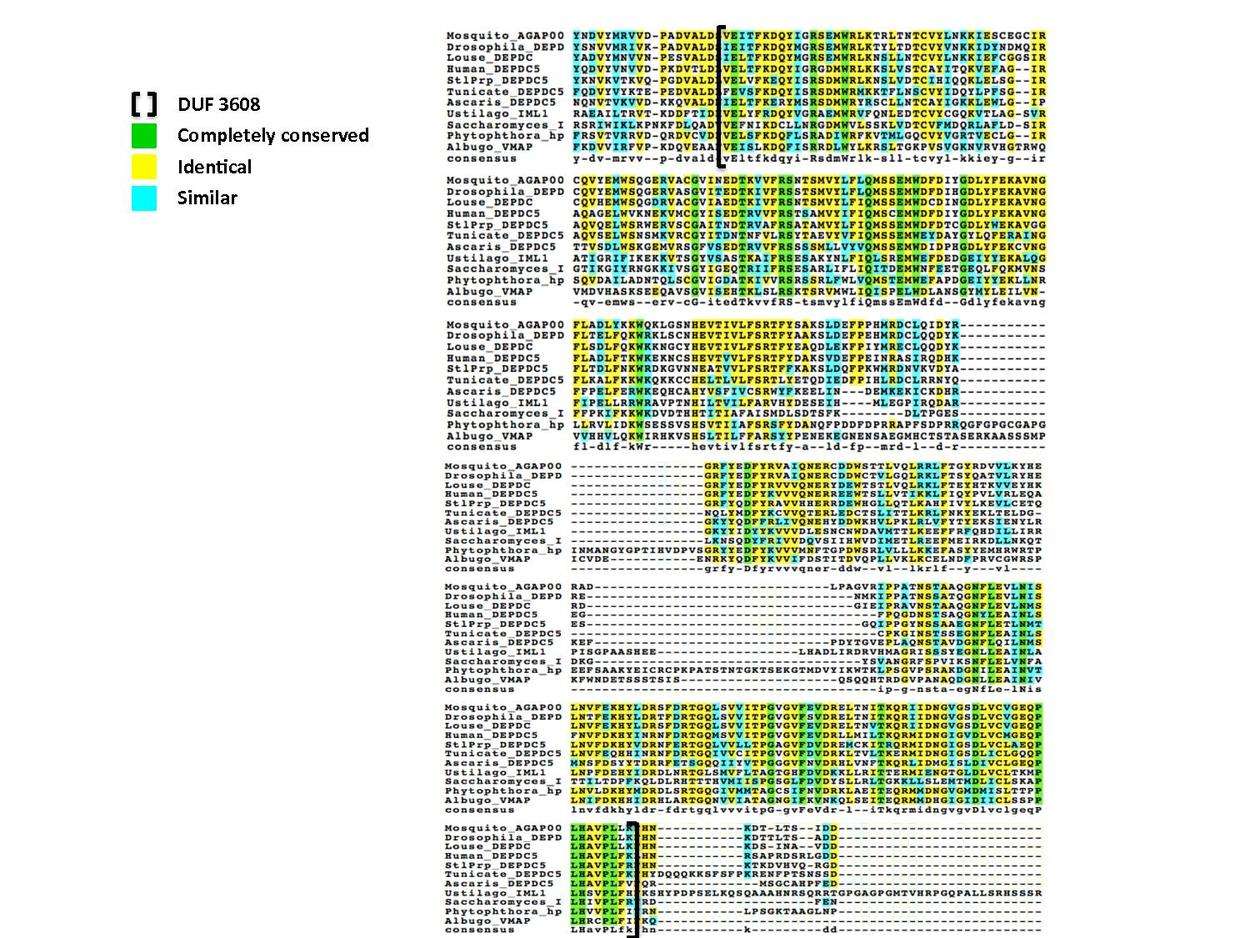
Paralogs
There are no known human DEPDC5 paralogs,[13] but there are 64 human proteins containing a homologous DEP domain.[16] There are also no identified paralogs for the yeast protein Iml1, the most distantly related ortholog of human DEPDC5.[13]
Expression
DEPDC5 expression has been characterized as ubiquitous in human tissue by RT-PCR analysis[17] and in DNA microarray studies as displayed in the chart below.[18]

One study on patients with hepatocellular carcinoma found higher DEPDC5 expression in tumor tissue than in non-tumor tissue.[4] Conversely, a homozygous deletion of three genes, one being DEPDC5, was found in two glioblastoma cases.[5] Other expression anomalies include zero expression in MDA-MB-231 breast cancer cell line[19] and low expression in P116 (ZAP70 negative) cell line.[20]
Post-translational Modifications
The following post-translational modifications were predicted with the proteomic tools compiled at ExPASy[21] and PhosphoSite Plus[22] for the human DEPDC5 protein.
| Post-translational Modification | Number/Loci | Source |
| Phosphorylation | 133/(Ser: 87 Thr: 23 Tyr: 23) | NetPhos |
| 6/S579, S582, S1499, Y1515, Y1519, Y1543 | PhosphoSite Plus | |
| Glycation | 29/5, 8, 13, 14, 28, 34, 56, 59, 64, 93, 131, 147, 229, 247, 256, 319, 436, 528, 609, 710, 862, 878, 1008, 1185, 1233, 1387, 1408, 1499, 1567, 1597 | NetGlycate |
| N-glycosylation site | 9/N201, N298, N311, N384, N684, N1157, N1377, N1444, N1529 | NetNGlyc |
| Sulfation | 3/Y397, Y459, Y462 | Sulfinator |
| Sumoylation | 2/K59, K147 | SUMOsp |
| Propeptide cleavage | 2/R1004-M1005, R1528-N1529 | ProP |
| O-glycosylation | 0 | NetOGlyc |
| C-mannosylation | 0 | NetCGlyc |
| Myristoylation | 0 | Myristoylation |
| Prenylation | 0 | PrePS |
| Acetylation | 0 | NetAcet |
Interaction
DEPDC5 may possibly interact with the proteasome subunit PSMA3 as evidenced by coimmunoprecipitation[23] and the transcription factor MYC.[24] DEPDC5 is in the "GATOR1" complex with Nprl2 and Nprl3.[25]
References
- ↑ "Diseases that are genetically associated with DEPDC5 view/edit references on wikidata".
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 3 Miki D, Ochi H, Hayes CN, et al. (August 2011). "Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers". Nat. Genet. 43 (8): 797–800. doi:10.1038/ng.876. PMID 21725309.
- 1 2 Seng TJ, Ichimura K, Liu L, Tingby O, Pearson DM, Collins VP (June 2005). "Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis". Genes Chromosomes Cancer. 43 (2): 181–93. doi:10.1002/gcc.20181. PMID 15770670.
- 1 2 "GeneCards: DEP domain containing 5".
- ↑ "AceView: Homo sapiens complex locus DEPDC5, encoding DEP domain containing 5".
- ↑ "SWISS-MODEL".
- ↑ "NCBI: CBLAST".
- ↑ Pan WJ, Pang SZ, Huang T, Guo HY, Wu D, Li L (August 2004). "Characterization of function of three domains in dishevelled-1: DEP domain is responsible for membrane translocation of dishevelled-1". Cell Res. 14 (4): 324–30. doi:10.1038/sj.cr.7290232. PMID 15353129.
- ↑ "Biology Workbench: PELE".
- ↑ Wu X, Tu BP (November 2011). "Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation". Mol. Biol. Cell. 22 (21): 4124–33. doi:10.1091/mbc.E11-06-0525. PMC 3204073
 . PMID 21900499.
. PMID 21900499. - 1 2 3 "NCBI".
- ↑ "TimeTree".
- ↑ "Biology Workbench: ClustalW".
- ↑ Civera C, Simon B, Stier G, Sattler M, Macias MJ (February 2005). "Structure and dynamics of the human pleckstrin DEP domain: distinct molecular features of a novel DEP domain subfamily". Proteins. 58 (2): 354–66. doi:10.1002/prot.20320. PMID 15573383.
- ↑ Ishikawa K, Nagase T, Suyama M, et al. (June 1998). "Prediction of the coding sequences of unidentified human genes. X. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro". DNA Res. 5 (3): 169–76. doi:10.1093/dnares/5.3.169. PMID 9734811.
- ↑ Johnson JM, Castle J, Garrett-Engele P, et al. (December 2003). "Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays". Science. 302 (5653): 2141–4. doi:10.1126/science.1090100. PMID 14684825.
- ↑ Cappellen D, Schlange T, Bauer M, Maurer F, Hynes NE (January 2007). "Novel c-MYC target genes mediate differential effects on cell proliferation and migration". EMBO Rep. 8 (1): 70–6. doi:10.1038/sj.embor.7400849. PMC 1796762
 . PMID 17159920.
. PMID 17159920. - ↑ Roose JP, Diehn M, Tomlinson MG, et al. (November 2003). "T cell receptor-independent basal signaling via Erk and Abl kinases suppresses RAG gene expression". PLoS Biol. 1 (2): E53. doi:10.1371/journal.pbio.0000053. PMC 261890
 . PMID 14624253.
. PMID 14624253. 
- ↑ "ExPASy Proteomic Tools: Post-translational modification prediction".
- ↑ "PhosphoSite Plus: DEPDC5".
- ↑ Lim J, Hao T, Shaw C, et al. (May 2006). "A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration". Cell. 125 (4): 801–14. doi:10.1016/j.cell.2006.03.032. PMID 16713569.
- ↑ Zeller KI, Zhao X, Lee CW, et al. (November 2006). "Global mapping of c-Myc binding sites and target gene networks in human B cells". Proc. Natl. Acad. Sci. U.S.A. 103 (47): 17834–9. doi:10.1073/pnas.0604129103. PMC 1635161
 . PMID 17093053.
. PMID 17093053. - ↑ Bar-Peled, L; Chantranupong, L; Cherniack, A. D.; Chen, W. W.; Ottina, K. A.; Grabiner, B. C.; Spear, E. D.; Carter, S. L.; Meyerson, M; Sabatini, D. M. (2013). "A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1". Science. 340 (6136): 1100–6. doi:10.1126/science.1232044. PMC 3728654
 . PMID 23723238.
. PMID 23723238.