Digital transcriptome subtraction
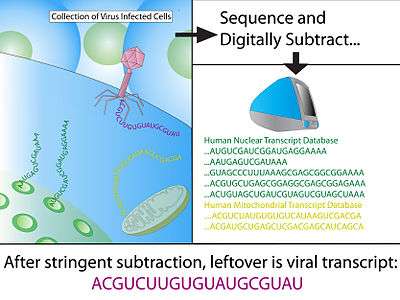
Digital transcriptome subtraction (DTS) is a bioinformatics method to detect the presence of novel pathogen transcripts through computational removal of the host sequences. DTS is the direct in silico analogue of the wet-lab approach Representational Difference Analysis (RDA), and is made possible by unbiased high-throughput sequencing and the availability of a high-quality, annotated reference genome of the host. The method specifically examines the etiological agent of infectious diseases and is best known for discovering Merkel cell polymavirus, the suspect causative agent in Merkel cell carcinoma.[1]
History
Using computational subtraction to discover novel pathogens was first proposed in 2002 by Meyerson et al.[2] using human expressed sequence tag (EST) datasets. In a proof of principle experiment, Meyerson et al. demonstrated that it was a feasible approach using Epstein-Barr virus-infected lymphocytes in post-transplant lymphoproliferative disorder (PTLD).[3]
In 2007, the term "Digital Transcriptome Subtraction" was coined by the Chang-Moore group,[4] and was used to discover Merkel cell polymavirus in Merkel cell carcinoma.[1]
Simultaneously to the MCV discovery, this approach was used to implicate a novel arenavirus as cause of fatality in a case where three patients died of similar illnesses shortly following organ transplantations from a single donor.[5]
Method
Construction of cDNA library
After treatment with DNase I to eliminate human genomic DNA, total RNA is extracted from primary infected tissue. Messenger RNA is then purified using an oligo-dT column that binds to the poly-A tail, a signal specifically found on transcribed genes. Using random hexamers priming, reverse transcriptase (RT) convert all mRNA into cDNA and cloned into bacterial vectors. Bacteria, usually E. coli, are then transformed using the cDNA vectors and selected using a marker, the collection of transformed clones is the cDNA library. This generates a snap-shot of tissue mRNA that is stable and can be sequenced at a later stage.
Sequencing and quality control
The cDNA library must be sequenced to great depth (i.e. number of clones sequenced) in order to detect a theoretical rare pathogen sequence (Table 1), especially if the foreign sequence is novel. Chang-Moore recommend a sequencing depth of 200,000 transcripts or greater using multiple sequencing platforms.[1]
| % Viral | 5,000 clones | 10,000 clones | 20,000 clones | 50,000 clones |
|---|---|---|---|---|
| 0.001% | 4.9% | 9.5% | 18.1% | 39.3% |
| 0.01% | 39.3% | 32.2% | 86.5% | 99.3% |
| 0.02% | 63.2% | 86.5% | 98.2% | >99.995% |
| 0.03% | 77.7% | 95.5% | 99.8% | >99.995% |
| 0.04% | 86.5% | 98.2% | >99.995% | >99.995% |
| 0.1% | 99.3% | >99.995% | >99.995% | >99.995% |
Stringent quality control are then applied to the raw sequences to minimize false-positive results. The initial quality screen uses several general parameters to exclude ambiguous sequences, leaving behind a dataset of high-fidelity (Hi-Fi) reads.
- Low Phred score cutoff is used to remove low-quality end sequences. Typically, a Phred score cutoff of 20 or 30 is used to ensure 99%-99.9% accuracy in each base-calling.
- Vector and adaptor removal.
- Low complexity - complexity score of a sequence reflects number of identical bases in a series (homo-polymers) such as poly-dT or poly-dA.
- Human repetitive DNA.
- Length - parameter is dependent on the optimized read length specific to the sequencing technology that was used.
- BLAST and exclude E. coli genome sequences.
BLAST to host genome
Using MEGABLAST, Hi-Fi reads are then matched to sequences in annotated databases and any positive matches are then subtracted from the dataset. Minimum hit length for a positive match of human sequence is typically 30 consecutive identical bases, which equates to a BLAST score of 60; generally, the remaining sequence is BLAST again with less stringent parameters to allow for slight mismatches (1 in 20 nucleotide). The vast majority of sequences (>99%) should be removed from the dataset at this stage.
Subtracted sequences typically include:
- Reference human transcriptome - eliminates any known human transcripts from expression library sets.
- Reference human genome - eliminates genes that have been missed by the annotation process and any contaminating genomic sequences during cDNA library construction.
- Mitochondrial DNA - mitochondrial DNA are highly abundant and polymorphic due to rapid mutation rate.
- Immunoglobulin region - The immunoglobulin loci is highly polymorphic and would otherwise yield false-positive due to poor alignment to the reference genome.
- Other vertebrate sequences
- Unannotated sequences
Analysis of "non-host" candidates
Alignment to pathogen databases
After stringent rounds of subtraction, the remaining sequences are clustered into non-redundant contigs and aligned to known pathogen sequences using low-stringency parameters. As pathogen genomes mutates quickly, nucleotide-nucleotide alignments, or blastn, is usually uninformative as it is possible to have mutations at certain bases without changing the amino acid residue due to codon degeneracy. Matching the in silico translated protein sequences of all 6 open reading frames to the amino acid sequence to annotated proteins, or blastx, is the preferred alignment method as it increases the likelihood of identifying a novel pathogen by matching to a related strain/species.[5] Experimental extension of candidate sequences might also be used at this stage to maximize chances of a positive match.[6]
De novo assembly
In cases where alignment to known pathogens is uninformative or ambiguous, contigs of candidate sequence can be used as templates for primer walking in primary infected tissue to generate the complete pathogen genome sequence.[1][5] As viral transcripts are exceedingly rare ratio tissue mRNA (10 transcripts in 1 million),[1] it is unlikely to generate a transcriptome based on the original candidate sequences alone due to low coverage.
Validation of pathogen
Once a putative pathogen has been identified in the high-throughput sequencing data, it is imperative to validate the presence of pathogen in infected patients using more sensitive techniques, such as:
- RT-PCR and derivative methods, including 3'- and 5'-RACE to confirm the existence of pathogen mRNA.
- Immunohistochemistry using antibodies to related pathogen to determine existence the pathogen in tissues.
- Serological tests to measure pathogen-specific antibody titer.
- Bacterial culture/viral culture, which is considered as the gold standard in laboratory diagnosis.
Applications
The primary application for DTS lies in identification of pathogenic viruses in cancer.[1][4] It can also be used to identify viral pathogens in non-cancer related disease.[5] Future clinical applications could include the use of DTS on a routine basis in individuals. DTS could also apply to agriculture, identifying pathogens that have an effect on output. Computation subtraction was already used in a metagenomics study that associated viral infection by IAPV with colony collapse disorder in honey bees.[7]
Advantages
- Requires no prior knowledge about pathogen sequence.[8]
- Can identify previously unassociated, potentially treatable pathogens.
- Uses already available molecular methods and resources.
Disadvantages
- Identifies the presence of pathogen but does not establish causal link to disease.[8] See Koch's postulate and Bradford-Hill criteria.
- Must have a highly reliable, complete reference transcriptome for the organism being studied.[8]
- Lack of foreign sequence identification cannot entirely exclude a pathogenic foreign body.[8]
References
- 1 2 3 4 5 6 Feng H, Shuda M, Chang Y, Moore PS (Jan 2008). "Clonal integration of a polyomavirus in human Merkel cell carcinoma.". Science. 5866. 319: 1096–1100. doi:10.1126/science.1152586. PMC 2740911
 . PMID 18202256.
. PMID 18202256. - 1 2 Weber G, Shendure J, Tanenbaum DM, Church GM, Meyerson M (Feb 2002). "Identification of foreign gene sequences by transcript filtering against the human genome.". Nat Genet. 2. 30: 141–142. doi:10.1038/ng818. PMID 11788827.
- 1 2 Xu Y, Stange-Thomann N, Weber G, Bo R, Dodge S, David RG, Foley K, Beheshti J, Harris NL, Birren B, Lander ES, Meyerson M (Mar 2003). "Pathogen discovery from human tissue by sequence-based computational subtraction.". Genomics. 3. 81: 329–335. doi:10.1016/S0888-7543(02)00043-5. PMID 12659816.
- 1 2 Feng H, Taylor JL, Benos PV, Newton R, Waddell K, Lucas SB, Chang Y, Moore PS (August 2007). "Human Transcriptome Subtraction by Using Short Sequence Tags To Search for Tumor Viruses in Conjunctival Carcinoma". J Virol. 20. 81: 11332–11340. doi:10.1128/JVI.00875-07. PMC 2045575
 . PMID 17686852.
. PMID 17686852. - 1 2 3 4 Palacios G, Druce J, Du L, Tran T, Birch C, Briese T, Conlan S, Quan PL, Hui J, Marshall J, Simons JF, Egholm M, Paddock CD, Shieh WJ, Goldsmith CS, Zaki SR, Catton M, Lipkin WI (Mar 2008). "A new arenavirus in a cluster of fatal transplant-associated diseases.". N Engl J Med. 10. 358: 991–998. doi:10.1056/NEJMoa073785. PMID 18256387.
- ↑ Chang Y, Moore PS. "New Pathogen Discovery: Digital Transcriptome Subtraction". Retrieved 1 March 2012.
- ↑ Cox-Foster DL, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA, Quan PL, Briese T, Hornig M, Geiser DM, Martinson V, vanEngelsdorp D, Kalkstein AL, Drysdale A, Hui J, Zhai J, Cui L, Hutchison SK, Simons JF, Egholm M, Pettis JS, Lipkin WI (Oct 2007). "A metagenomic survey of microbes in honey bee colony collapse disorder.". Science. 5848. 318: 283–287. doi:10.1126/science.1146498. PMID 17823314.
- 1 2 3 4 MacConaill L, Meyerson M (Apr 2008). "Adding pathogens by genomic subtraction.". Nat Genet. 4. 40: 380–382. doi:10.1038/ng0408-380. PMID 18368124.