Ectoderm specification
In Xenopus laevis, the specification of the three germ layers (endoderm, mesoderm and ectoderm) occurs at the blastula stage.[1] Great efforts have been made to determine the factors that specify the endoderm and mesoderm. On the other hand, only a few examples of genes that are required for ectoderm specification have been described in the last decade. The first molecule identified to be required for the specification of ectoderm was the ubiquitin ligase Ectodermin (Ecto, TIF1-γ, TRIM33); later, it was found that the deubiquitinating enzyme, FAM/USP9x, is able to overcome the effects of ubiquitination made by Ectodermin in Smad4 (Dupont et al., 2009). Two transcription factors have been proposed to control gene expression of ectodermal specific genes: POU91/Oct3/4[2] and FoxIe1/Xema.[3][4] A new factor specific for the ectoderm, XFDL156, has shown to be essential for suppression of mesoderm differentiation from pluripotent cells.[5]
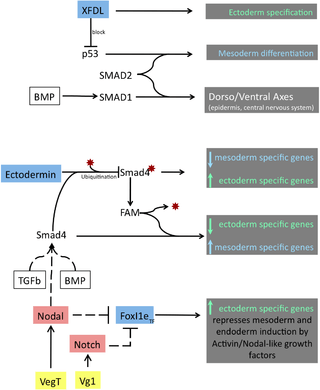
Ectodermin and FAM
Biological role of Ectodermin and FAM
The protein Ectodermin, firstly identified in Xenopus embryos, promotes ectodermal fate and suppresses the mesoderm formation mediated by the signaling of Transforming Growth Factor β (TGFβ) and Bone Morphogenic Proteins (BMP), members of the TGFβ-superfamily.[6] When the TGFβ ligands bind to TGFβ receptors, they cause the activation of the signal transducers R-Smads (Smad2, Smad3). Smad4 forms a complex with activated R-Smads and activates transcription of specific genes in response to TGFβ signal. The BMP pathway transmits its signals in a similar way but through other types of R-Smads (Smad1, Smad5 and Smad8). The transcription factor Smad4 is the only common mediator shared between both TGFβ and the BMP pathways.[7] During ectoderm specification, the function of Smad4 is regulated by ubiquitination and deubiquitination made by Ectodermin and FAM (acronym of Fat acets in mammals) respectively. The ubiquitination state of Smad4 will determine if it is be able to respond to signals derived from TGFβ and BMP.[6][8] The equilibrium of the activity, localization and timing of TGFβ and BMP transducers, Smad4, FAM and of Ectodermin should be achieved in order to be able to modulate the gene expression of genes required for germ layer formation.
Identification of Ectodermin and FAM
A cDNA library from the blastula stage of a frog embryo was cloned into RNA expression plasmids to generate synthetic mRNA. The mRNA was then injected into several Xenopus embryos at a four-cell stage and looked in early blastula embryos for an expansion of the region of the ectodermal marker Sox2 and diminution of the expression of the mesodermal marker Xbra. Ectodermin was one out of 50 clones to present this phenotype when injected into embryos.[6] The identification of FAM was done through a siRNA screen to find deubiquitinases that regulate the response to TGFβ.
Ectodermin and FAM localization
Ectodermin mRNA is maternally deposited in the animal pole of the egg. In the early blastula stage of the embryo, Ectodermin mRNA and protein forms a gradient that goes from the animal pole (highest concentration) down to the marginal zone (lowest concentration) to prevent TGFβ and nodal signals that induce mesoderm originating from the vegetal pole. Ectodermin mRNA is enriched in the dorsal side of the embryo, and at the end of this stage the expression gradually disappears.[6] Smad4 is ubiquitinated by Ectodermin in the nucleus and exported to the cytoplasm where it can be deubiquitinated by FAM; this way Smad4 can be recycled and be functional again. Although there is no expression profile of FAM in early embryos in Xenopus, in the zebra fish, FAM homolog is expressed ubiquitously at a two-cell stage but as development proceeds then its only expressed in the cephalic central nervous system.[9]
Ectodermin and FAM functions
Ectodermin is a ubiquitin E3 ligase that inhibits the TGFβ and the BMP signaling pathways by inhibition of Smad4 via ubiquitination of Lysine 519 and also though direct binding to phospho-Smad2.[6][8] Injection of Ecto mRNA in the marginal zone leads to an expansion of the early ectodermal marker, Sox2, and a reduction of mesodermal markers (Xbra, Eomes, Vent-1, Mix-1 and Mixer). The opposite happens in knockdown experiments of Ectodermin by using a morpholino strategy; embryos become more sensitive to Activin response, they show an increase and expansion of the expression of mesodermal specific genes and down-regulate the expression of neural plate and epidermis marker (Sox2 and cytokeratin respectively). In line with a RING-finger dependent ubiquitin-ligase activity of Ectodermin, an Ecto RING-finger mutant (C97A/C100A) is inactive in gain-of-function.[6] Gain-of-function of FAM increases the responses from BMP and TGFβ and its loss-of-function by mutation in a critical residue for its activity caused inhibition of TGFβ response.
Conservation of Ectodermin and FAM in other species
The molecular function of human ectodermin to act as a negative regulator of Smad4 suggests that this specific function is conserved among the vertebrate lineage.[6] The sequence identity between FAM homologs is higher than 90% when comparing the homologs of Xenopus, zebrafish, mouse, and human, suggesting that this might also be conserved among other organisms.[9] Indeed, knockout gene inactivation in mouse embryos showed that the function of ectodermin as inhibitor of TGF-beta signaling is conserved.[10] Embryos lacking of ectodermin show defective development of the anterior visceral endoderm (AVE), which is the first tissue that is induced by TGF-beta signals in mouse embryos; in accordance with loss of an inhibitor, ectodermin-/- embryos showed enlarged AVE induction. As the AVE is a natural source of secreted TGF-beta antagonists, this primary AVE expansion caused secondarily, at later stages, an inhibition of extracellular TGF-beta ligands, resulting in embryos lacking of mesoderm development. This model was confirmed by the finding that ectodermin-/- embryos were rescued to wild type (normal AVE, normal mesoderm development) by lowering the genetic dosage of the main TGF-beta ligand of the emrbyo, Nodal. Further supporting a role as TGF-beta inhibitor, tissue-selective deletion of ectodermin from the epiblast (from which the mesoderm, but not the AVE, derive) left the AVE untouched but caused this time an expansion of anterior mesodermal fates, indicative of increased responsiveness to TGF-beta signals. Collectively, these data confirmed with genetic tools a cell-autonomous role for ectodermin as inhibitor of Smad4 responses previously identified in Xenopus embryos and human cell lines.
FOXI1e
Biological role of FOXI1e
During early in development in Xenopus, the transcription factor FoxI1e/Xema activates epidermal differentiation and represses endoderm and mesoderm specific genes in animal caps (Suri et al., 2005). It is suggested that FoxI1e is active before the ectoderm differentiates into epidermis and the central nervous system.
Identification of FoxI1e
Mir et al., 2005 identified FoxI1e (Xema) by selecting genes that were down-regulated under mesoderm-inducing signals in the ectoderm compared to vegetal region of an early blastula embryo. Also, high expression of this gene was observed in animal caps in embryos that lack VegT compared to wild type.
Localization in the cell
FoxI1e mRNA is expressed zygotically (stage 8.5) and reaches higher level of expression early in gastrulation and maintains that level in neurula, tailbud until early tadpole stages.[4] FoxI1e has a peculiar mosaic expression pattern, it is expressed first in the dorsal ectoderm and while gastrula progresses, the expression goes through the ventral side and its expression is down-regulated in the dorsal side when the neural plate is forming.[11] FoxI1e is dependent on BMP signals in the neurula stage, limiting the localization of FoxI1e to the ventral side of the ectoderm.
Protein function and regulation
FoxI1e/Xema belongs to the FoxI1 class of fork head transcription factor family, known to participate in mesoderm formation, eye development[12] and ventral head specification.[13] It has been proposed that Notch and/or NODAL, expressed in the vegetal/mesoderm region of the early blastula embryo, could potentially be the inhibitors of FoxI1e.[3][11]
Loss and gain-of-function
Inhibition of FoxI1e mRNA maturation by a splice-blocking morpholino shows malformations in the development of epidermis and pervious system and down-regulates of ectoderm specific genes, whereas FoxI1e over-expression inhibits the formation of mesoderm and endoderm. Vegetal structures form late blastula masses that normally would give rise to endoderm and mesoderm, when injected with FoxI1e mRNA, they are able to express ectodermal specific markers (pan-ectodermal E-cadherin, epithelial cytokeratin, neural crest marker Slug and neural marker Sox-2) while endodermal markers (endodermin, Xsox17a) decreased in expression.[3][4]
XFDL156
Biological role of XFDL156
The p53 protein binds to the promoters of early mesodermal genes.[14] p53 is maternally deposited transcript that forms a transcriptional factor complex with Smad2 and drives the expression of genes involved in mesoderm induction and activation of TGFβ target genes.[15] The zinc (Zn)-finger nuclear protein XFDL159, expressed in the animal cap, acts as an ectoderm factor their specifies the ectoderm by inhibiting p53 from activating genes for mesoderm differentiation.[5]
Identification of XFDL156
Construction of a cDNA library from animal caps at a stage of 11.5, cloned into expression vector and generated mRNA. The synthetic RNA was then injected into embryos and the animal caps of these collected embryos were obtained and submitted to activin treatment. Xbra was recovered by selecting the clone that represses the mesodermal marker Xbra.[5]
Localization of XFDL156
Since XFDL156 is a factor that interacts with p53, the localization of this protein is in the nucleus (Sasai et al., 2008). The mRNA of XFDL156 is maternally deposited and then expressed zygotically. The gene expression timeline shows a higher level of expression at early gastrula and a half decrease in expression at mid-gastrula and by the stage 20 the expression fades.[5]
Protein function of XFDL156
XFDR zinc finger binds to the regulatory region of p53 located at the C-terminal domain and its expression is not affected by the presence of activin, FoxI1e or XLPOU91 transcription factors.
Loss and gain-of-function in XFDL156
Loss-of-function by morpholino, causes incorrect mesodermal differentiation in the ectoedermal regions; this is caused by the desuppression of mesodermal markers (Xbra, VegT and Mix.2). Gain-of-functions causes decrease in expression of mesodermal markers.[5]
Conservation of XFDL homologs in other species
The human and mouse homologs of XFDR156 are able to complement XFDR function of interacting with p53 and inhibiting it to act as a transcription factor.[5]
-
Ectoderm induction in early blastula in Xenopus embryo.
References
- ↑ Heasman, J., Quarmby, J., and Wylie, C.C. (1984). The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev Biol 105, 458-469.
- ↑ Snir, M., Ofir, R., Elias, S., and Frank, D. (2006). Xenopus laevis POU91 protein, an Oct3/4 homologue, regulates competence transitions from mesoderm to neural cell fates. EMBO J 25, 3664-3674.
- 1 2 3 Mir, A., Kofron, M., Zorn, A.M., Bajzer, M., Haque, M., Heasman, J., and Wylie, C.C. (2007). FoxI1e activates ectoderm formation and controls cell position in the Xenopus blastula. Development 134, 779-788.
- 1 2 3 Suri, C., Haremaki, T., and Weinstein, D.C. (2005). Xema, a foxi-class gene expressed in the gastrula stage Xenopus ectoderm, is required for the suppression of mesendoderm. Development 132, 2733-2742.
- 1 2 3 4 5 6 Sasai, N., Yakura, R., Kamiya, D., Nakazawa, Y., and Sasai, Y. (2008). Ectodermal factor restricts mesoderm differentiation by inhibiting p53. Cell 133, 878-890.
- 1 2 3 4 5 6 7 Dupont, S., Zacchigna, L., Cordenonsi, M., Soligo, S., Adorno, M., Rugge, M., and Piccolo, S. (2005). Germ-layer specification and control of cell growth by Ectodermin, a Smad4 ubiquitin ligase. Cell 121, 87-99.
- ↑ Siegel, P.M., and Massague, J. (2003). Cytostatic and apoptotic actions of TGFβeta in homeostasis and cancer. Nat Rev Cancer 3, 807-821.
- 1 2 Dupont, S., Mamidi, A., Cordenonsi, M., Montagner, M., Zacchigna, L., Adorno, M., Martello, G., Stinchfield, M.J., Soligo, S., Morsut, L., et al. (2009). FAM/USP9x, a deubiquitinating enzyme essential for TGFβeta signaling, controls Smad4 monoubiquitination. Cell 136, 123-135.
- 1 2 Khut, P.Y., Tucker, B., Lardelli, M., and Wood, S.A. (2007). Evolutionary and expression analysis of the zebrafish deubiquitylating enzyme, usp9. Zebrafish 4, 95-101.
- ↑ Morsut L, Yan KP, Enzo E, Aragona M, Soligo SM, Wendling O, Mark M, Khetchoumian K, Bressan G, Chambon P, Dupont S, Losson R, Piccolo S (2010). "Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo". Development. 137 (15): 2571–8. doi:10.1242/dev.053801. PMID 20573697.
- 1 2 Mir, A., Kofron, M., Heasman, J., Mogle, M., Lang, S., Birsoy, B., and Wylie, C. (2008). Long- and short-range signals control the dynamic expression of an animal hemisphere-specific gene in Xenopus. Dev Biol 315, 161-172.
- ↑ Pohl, B.S., Rossner, A., and Knochel, W. (2005). The Fox gene family in Xenopus laevis:FoxI2, FoxM1 and FoxP1 in early development. Int J Dev Biol 49, 53-58.
- ↑ Matsuo-Takasaki, M., Matsumura, M., and Sasai, Y. (2005). An essential role of Xenopus Foxi1a for ventral specification of the cephalic ectoderm during gastrulation. Development 132, 3885-3894.
- ↑ Cordenonsi, M., Dupont, S., Maretto, S., Insinga, A., Imbriano, C., and Piccolo, S. (2003). Links between tumor suppressors: p53 is required for TGFβeta gene responses by cooperating with Smads. Cell 113, 301-314.
- ↑ Takebayashi-Suzuki, K., Funami, J., Tokumori, D., Saito, A., Watabe, T., Miyazono, K., Kanda, A., and Suzuki, A. (2003). Interplay between the tumor suppressor p53 and TGF beta signaling shapes embryonic body axes in Xenopus. Development 130, 3929-3939.