HIST1H2BG
Histone H2B type 1-C/E/F/G/I is a protein that in humans is encoded by the HIST1H2BG gene.[3][4][5]
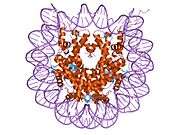
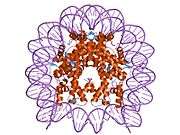
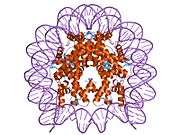
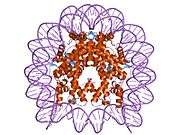
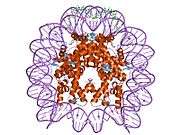
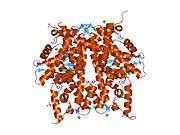
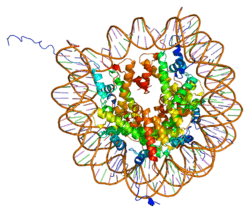
Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a member of the histone H2B family. Transcripts from this gene lack polyA tails; instead, they contain a palindromic termination element. This gene is found in the large histone gene cluster on chromosome 6p22-p21.3.[5]
References
Further reading
- Ohe Y, Hayashi H, Iwai K (1979). "Human spleen histone H2B. Isolation and amino acid sequence.". J. Biochem. 85 (2): 615–24. PMID 422550.
- Frohm M, Gunne H, Bergman AC, et al. (1996). "Biochemical and antibacterial analysis of human wound and blister fluid.". Eur. J. Biochem. 237 (1): 86–92. doi:10.1111/j.1432-1033.1996.0086n.x. PMID 8620898.
- Díaz-Jullien C, Pérez-Estévez A, Covelo G, Freire M (1996). "Prothymosin alpha binds histones in vitro and shows activity in nucleosome assembly assay.". Biochim. Biophys. Acta. 1296 (2): 219–27. doi:10.1016/0167-4838(96)00072-6. PMID 8814229.
- Albig W, Kioschis P, Poustka A, et al. (1997). "Human histone gene organization: nonregular arrangement within a large cluster.". Genomics. 40 (2): 314–22. doi:10.1006/geno.1996.4592. PMID 9119399.
- Albig W, Doenecke D (1998). "The human histone gene cluster at the D6S105 locus.". Hum. Genet. 101 (3): 284–94. doi:10.1007/s004390050630. PMID 9439656.
- El Kharroubi A, Piras G, Zensen R, Martin MA (1998). "Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter.". Mol. Cell. Biol. 18 (5): 2535–44. doi:10.1128/mcb.18.5.2535. PMC 110633
 . PMID 9566873.
. PMID 9566873.
- Piredda L, Farrace MG, Lo Bello M, et al. (1999). "Identification of 'tissue' transglutaminase binding proteins in neural cells committed to apoptosis.". FASEB J. 13 (2): 355–64. PMID 9973324.
- Deng L, de la Fuente C, Fu P, et al. (2001). "Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones.". Virology. 277 (2): 278–95. doi:10.1006/viro.2000.0593. PMID 11080476.
- Deng L, Wang D, de la Fuente C, et al. (2001). "Enhancement of the p300 HAT activity by HIV-1 Tat on chromatin DNA.". Virology. 289 (2): 312–26. doi:10.1006/viro.2001.1129. PMID 11689053.
- Andersen JS, Lyon CE, Fox AH, et al. (2002). "Directed proteomic analysis of the human nucleolus.". Curr. Biol. 12 (1): 1–11. doi:10.1016/S0960-9822(01)00650-9. PMID 11790298.
- Kim HS, Cho JH, Park HW, et al. (2002). "Endotoxin-neutralizing antimicrobial proteins of the human placenta.". J. Immunol. 168 (5): 2356–64. doi:10.4049/jimmunol.168.5.2356. PMID 11859126.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241
 . PMID 12477932.
. PMID 12477932.
- Cheung WL, Ajiro K, Samejima K, et al. (2003). "Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase.". Cell. 113 (4): 507–17. doi:10.1016/S0092-8674(03)00355-6. PMID 12757711.
- Tollin M, Bergman P, Svenberg T, et al. (2004). "Antimicrobial peptides in the first line defence of human colon mucosa.". Peptides. 24 (4): 523–30. doi:10.1016/S0196-9781(03)00114-1. PMID 12860195.
- Mungall AJ, Palmer SA, Sims SK, et al. (2003). "The DNA sequence and analysis of human chromosome 6.". Nature. 425 (6960): 805–11. doi:10.1038/nature02055. PMID 14574404.
- Coleman MA, Miller KA, Beernink PT, et al. (2004). "Identification of chromatin-related protein interactions using protein microarrays.". Proteomics. 3 (11): 2101–7. doi:10.1002/pmic.200300593. PMID 14595808.
- Lusic M, Marcello A, Cereseto A, Giacca M (2004). "Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter.". EMBO J. 22 (24): 6550–61. doi:10.1093/emboj/cdg631. PMC 291826
 . PMID 14657027.
. PMID 14657027.
- Howell SJ, Wilk D, Yadav SP, Bevins CL (2004). "Antimicrobial polypeptides of the human colonic epithelium.". Peptides. 24 (11): 1763–70. doi:10.1016/j.peptides.2003.07.028. PMID 15019208.
PDB gallery |
|---|
|
| 1aoi: COMPLEX BETWEEN NUCLEOSOME CORE PARTICLE (H3,H4,H2A,H2B) AND 146 BP LONG DNA FRAGMENT |
| 1eqz: X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION |
| 1f66: 2.6 A CRYSTAL STRUCTURE OF A NUCLEOSOME CORE PARTICLE CONTAINING THE VARIANT HISTONE H2A.Z |
| 1hq3: CRYSTAL STRUCTURE OF THE HISTONE-CORE-OCTAMER IN KCL/PHOSPHATE |
| 1kx3: X-Ray Structure of the Nucleosome Core Particle, NCP146, at 2.0 A Resolution |
| 1kx4: X-Ray Structure of the Nucleosome Core Particle, NCP146b, at 2.6 A Resolution |
| 1kx5: X-Ray Structure of the Nucleosome Core Particle, NCP147, at 1.9 A Resolution |
| 1m18: LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA |
| 1m19: LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA |
| 1m1a: LIGAND BINDING ALTERS THE STRUCTURE AND DYNAMICS OF NUCLEOSOMAL DNA |
| 1p34: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3a: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3b: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3f: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3g: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3i: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3k: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3l: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3m: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3o: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1p3p: Crystallographic Studies of Nucleosome Core Particles containing Histone 'Sin' Mutants |
| 1s32: Molecular Recognition of the Nucleosomal 'Supergroove' |
| 1tzy: Crystal Structure of the Core-Histone Octamer to 1.90 Angstrom Resolution |
| 1u35: Crystal structure of the nucleosome core particle containing the histone domain of macroH2A |
| 1zbb: Structure of the 4_601_167 Tetranucleosome |
| 1zla: X-ray Structure of a Kaposi's sarcoma herpesvirus LANA peptide bound to the nucleosomal core |
| 2aro: Crystal Structure Of The Native Histone Octamer To 2.1 Angstrom Resolution, Crystalised In The Presence Of S-Nitrosoglutathione |
| 2cv5: Crystal structure of human nucleosome core particle |
| 2f8n: 2.9 Angstrom X-ray structure of hybrid macroH2A nucleosomes |
| 2fj7: Crystal structure of Nucleosome Core Particle Containing a Poly (dA.dT) Sequence Element |
| 2hio: HISTONE OCTAMER (CHICKEN), CHROMOSOMAL PROTEIN |
| 2nzd: Nucleosome core particle containing 145 bp of DNA |
|
|
 . PMID 9566873.
. PMID 9566873. . PMID 12477932.
. PMID 12477932. . PMID 14657027.
. PMID 14657027.