Resistance mutation
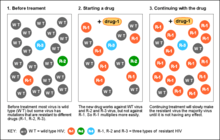
A resistance mutation is a mutation in a virus gene that allows the virus to become resistant to treatment with a particular antiviral drug. The term was first used in the management of HIV, the first virus in which genome sequencing was routinely used to look for drug resistance. At the time of infection, a virus will infect and begin to replicate within a preliminary cell. As subsequent cells are infected, random mutations will occur in the viral genome.[1] When these mutations begin to accumulate, antiviral methods will kill the wild type strain, but will not be able to kill one or many mutated forms of the original virus. At this point a resistance mutation has occurred because the new strain of virus is now resistant to the antiviral treatment that would have killed the original virus.[1] Resistance mutations are evident and widely studied in HIV due to its high rate of mutation and prevalence in the general population. Resistance mutation is now studied in bacteriology and parasitology.
Mechanisms
Resistance mutations can occur through several mechanisms from single nucleotide substitutions to combinations of amino acid substitutions, deletions and insertions.[1] Over time, these new genetic lines will persist if they become resistant to treatments being used against them. It has been shown that pathogens will favor and become more resistant to treatment in common host genotypes through frequency-dependent selection.[2] Further, strict adherence to a retroviral regimen correlates to a strong decrease in retroviral resistance mutations.[3] There are five classes of drug that are used to treat HIV infection, and resistance mutations can effect the efficacy of these treatments as well.
- Entry inhibitors block the ability of HIV to enter its target cells. HIV must bind to a CD4 receptor on a T cell or the CCR5/CXCR4 co-receptors to enter the cell. They can also block the fusion of the viral and cell membranes. Entry inhibitors modify protein residues on the membrane of the T cell to make it incompatible with the HIV, thus blocking insertion of the viral genome.[4]
- Nucleoside reverse transcriptase inhibitors (NRTI) are phosphorylated by cellular enzymes rendering it an active NRTI triphosphate stage. When the NRTIs are activated, they being to complete with cellular nucleosides that would be incorporated during reverse transcription by HIV. When the NRTIs are incorporated into the strand being transcribed, it causes the reverse transcription to be stopped.[5] This inhibits HIV from incorporating its genome into the host cell.
- Non-nucleoside reverse transcriptase inhibitors (NNRTI) inhibit reverse transcriptase by binding to an allosteric site on it. Cessation of a non-nucleoside reverse transcriptase inhibitor regimen poses a significant danger of selection for a drug-resistant form of HIV-1.[6] A K103N mutation in HIV-1 causes the hydrophobic pocket in which the NNRTI binds to inhibit enzyme activity in reverse transcriptase to close by means of a hydrogen bond. Without access to the allosteric site, the NNRTI will not be effective.[1]
- Integrase inhibitors prevent the HIV integrase enzyme from incorporating its viral genome into the host chromosome. The enzyme contains a highly conserved region called the catalytic core, which is essential for binding to the virally encoded DNA. Integrase inhibitors block the ability of the catalytic core to form covalent bonds with the phosphodiester backbone of DNA.[7] As a result, the viral DNA cannot be incorporated into the host genome.
- Protease Inhibitors cause the immature formation of two essential precursor proteins known as Gag and Gagpol in HIV. These proteins are essential in maintaining viral structure and for enzyme creation. Protease inhibitors bind to the active site of the viral protease, which prevents the cleavage of the Gag and Gagpol proteins. As a result, non-infections viral particles are formed.[8]
In Other Viruses
Resistance mutations are found and become problematic in many viruses other than HIV. Notable examples of such viruses include the herpes simplex virus and hepatitis B virus.[9] In the herpes virus, drugs mainly target the viral DNA polymerase. As a result, mutations in the viral DNA polymerase that make it resistant to these drugs are selected for, which ultimately can cause complete resistance of the treatment. In hepatitis B, nucleoside and nucleotide analogs are used to cause early termination of viral transcription. Mutations in viral reverse transcriptase can cause the enzyme to not incorporate these nonfunctional analogs, in favor for their natural counterparts. If this mutation occurs, transcription will not be halted, and viral proteins can be created.
Research Applications
- A study of hepatitis B in a Chinese population investigated the relationship of resistance mutations in the virus to its replication and genotype. It helped to gain insight into common mutations and their patterns. Patients undergoing treatment with a nucleoside drug were selected and fluorescence PCR and Sanger sequencing was used to sequence the viral genome. These sequences were compared against known hepatitis B sequences from Genbank so that the patterns of resistance could be analyzed. It was shown that many patients had a single mutation in common and many were also completely resist to a common nucleoside drug. In addition to gaining insight into mutation patterns, this study highlights the importance of using multiple types of drugs to treat chronic viral conditions.[10]
- An important part of treating HIV is patient-specific tailoring of drugs. A study identified the most common resistance mutations so that point of care tests could determine the resistance mutations of a patient, thereby allowing their provider to tailor their treatment. This point of care test would most likely be an inexpensive assay, which could significantly improve HIV treatment in impoverished areas. The researchers used datasets of the sequences of thousands of individuals around the world to identify the most common resistance mutations to NRTI, NNRTI, and protease inhibitors. Two NRTI mutations and four NNRTI mutations were commonly found, which can now be tested for using an inexpensive point-of-care.[11] This will significantly improve care quality and effectiveness, especially in impoverished areas.
References
- 1 2 3 4 Clavel, François. "Mechanisms of HIV Drug Resistance: A Primer" (PDF). Physicians' Research Network. Physicians Research Network Inc. Retrieved 6 February 2016.
- ↑ Bangham, Jenny; Obbard, Darren; Kim, Kang-Wook; Haddril, Penelope; Jiggins, Francis (22 August 2007). "The Age and Evolution of an Antiviral Resistance Mutation in Drosophila melanogaster". Proceedings: Biological Sciences. 274 (1621): 2027–2034. doi:10.1098/rspb.2007.0611.
- ↑ Harrigan, P.; Hogg, Robert; Dong, Winnie; Yip, Benita; Wynhoven, Brian; Woodward, Justin; Broome, Chanson; Broome, Zabrina; Mo, Theresa; Alexander, Chris; Montaner, Julio (1 February 2005). "Predictors of HIV Drug-Resistance Mutations in a Large Antiretroviral-Naive Cohort Initiating Triple Antiretroviral Therapy". The Journal of Infection Diseases. 151 (3): 339–347.
- ↑ Briz, Verónica; Poveda, Eva; Soriano, Vincent (2006-04-01). "HIV entry inhibitors: mechanisms of action and resistance pathways". The Journal of Antimicrobial Chemotherapy. 57 (4): 619–627. doi:10.1093/jac/dkl027. ISSN 0305-7453. PMID 16464888.
- ↑ Maga, Giovanni; Radi, Marco; Gerard, Marie-Aline; Botta, Maurizio; Ennifar, Eric (2010-03-30). "HIV-1 RT Inhibitors with a Novel Mechanism of Action: NNRTIs that Compete with the Nucleotide Substrate". Viruses. 2 (4): 880–899. doi:10.3390/v2040880. ISSN 1999-4915. PMC 3185657
 . PMID 21994659.
. PMID 21994659. - ↑ Hare, C. Bradley; Mellors, John; Krambrink, Amy; Su, Zhaohui; Skiest, Daniel; Margolis, David M.; Patel, Sheran S.; Barnas, Douglas; Frenkel, Lisa; Coombs, Robert W.; Aweeka, Francesca; Morse, Gene D.; Haas, David W.; Boltz, Valerie; Palmer, Sarah; Coffin, John; Havlir, Diane V. (August 2008). "Detection of Nonnucleoside Reverse‐Transcriptase Inhibitor–Resistant HIV‐1 after Discontinuation of Virologically Suppressive Antiretroviral Therapy". Clinical Infectious Diseases. 47 (3): 421–424. doi:10.1086/589867.
- ↑ Hicks, Charles; Gulick, Roy M. (2009-04-01). "Raltegravir: The First HIV Type 1 Integrase Inhibitor". Clinical Infectious Diseases. 48 (7): 931–939. doi:10.1086/597290. ISSN 1058-4838. PMID 19231980.
- ↑ van Maarseveen, Noortje; Boucher, Charles (2006-01-01). Geretti, Anna Maria, ed. Resistance to protease inhibitors. London: Mediscript. ISBN 9780955166907. PMID 21249774.
- ↑ Strasfeld, Lynne; Chou, Sunwen (2010-06-01). "Antiviral Drug Resistance: Mechanisms and Clinical Implications". Infectious disease clinics of North America. 24 (2): 413–437. doi:10.1016/j.idc.2010.01.001. ISSN 0891-5520. PMC 2871161
 . PMID 20466277.
. PMID 20466277. - ↑ He, Xiaohong; Wang, Fang; Huang, Bin; Chen, Peisong; Zhong, Liangying (2015-06-15). "Detection and analysis of resistance mutations of hepatitis B virus". International Journal of Clinical and Experimental Medicine. 8 (6): 9630–9639. ISSN 1940-5901. PMC 4538069
 . PMID 26309637.
. PMID 26309637. - ↑ Rhee, Soo-Yon; Jordan, Michael R.; Raizes, Elliot; Chua, Arlene; Parkin, Neil; Kantor, Rami; Zyl, Gert U. Van; Mukui, Irene; Hosseinipour, Mina C. "HIV-1 Drug Resistance Mutations: Potential Applications for Point-of-Care Genotypic Resistance Testing". PLOS ONE. 10 (12). doi:10.1371/journal.pone.0145772. PMC 4696791
 . PMID 26717411.
. PMID 26717411.
External links
- HIV drug resistance mutations figures
- FDA-Approved HIV Medicines
- CDC Explanation of Influenza Resistance Mutations