Frederic M. Richards
| Frederic Middlebrook Richards | |
|---|---|
 Fred Richards | |
| Born |
August 19, 1925 New York, New York |
| Died |
January 11, 2009 (aged 83) Guilford, Connecticut[1] |
| Residence | Guilford, Connecticut |
| Citizenship | United States |
| Fields | Biophysics, Biochemistry |
| Institutions | Yale University |
| Alma mater | MIT B.S. 1948, Harvard University Ph.D 1952 |
| Thesis | Studies on the density and composition of some protein crystals, including a preliminary investigation of the crystal structure of zinc diglycinate monohydrate[2] (1952) |
| Doctoral advisor | Barbara Low[3] |
| Other academic advisors |
Edwin Joseph Cohn Kaj Ulrik Linderstrøm-Lang[3] |
| Doctoral students | Jorge Allende[3] |
| Other notable students |
Cyrus Chothia Louise Johnson Wendell Lim[3] |
| Known for | Solving the 3rd-ever crystal structure of a protein – ribonuclease S; defining solvent-accessible surface |
| Influences | Christian B. Anfinsen, Hans T. Clarke |
| Influenced | Thomas A. Steitz, Jane S. Richardson |
| Notable awards |
National Academy of Sciences American Academy of Arts and Sciences Guggenheim Fellowship |
| Spouse | Heidi Clarke Richards; Sarah (Sally) Wheatland Richards |
Frederic Middlebrook Richards (August 19, 1925 – January 11, 2009), commonly referred to as Fred Richards, was an American biochemist and biophysicist known for solving the pioneering crystal structure of the ribonuclease S enzyme in 1967 and for defining the concept of solvent-accessible surface. He contributed many key experimental and theoretical results and developed new methods, garnering over 20,000 journal citations in several quite distinct research areas. In addition to the protein crystallography and biochemistry of ribonuclease S, these included solvent accessibility and internal packing of proteins, the first side-chain rotamer library, high-pressure crystallography, new types of chemical tags such as biotin/avidin, the nuclear magnetic resonance (NMR) chemical shift index, and structural and biophysical characterization of the effects of mutations.
Richards spent his entire academic research career at Yale University, where he became Sterling Professor of Molecular Biophysics and Biochemistry in the department that he created and chaired, "one of the major centers in the world for the study of biophysics and structural biology".[4] He was elected to the National Academy of Sciences USA and the American Academy of Arts and Sciences, and received many other scientific awards. He served as head of the Jane Coffin Childs Memorial Fund for Medical Research and was elected as president both of the American Society for Biochemistry and Molecular Biology (ASBMB) and of the Biophysical Society.

Personal biography
Richards was born on August 19, 1925 in New York City to George H. Richards and Marianna Middlebrook Richards. Both parents were from old New England families who had settled in Fairfield and New London, Connecticut in the 1600s. The family usually spent summers in Connecticut, giving Richards an early affinity for the area which continued through his career at Yale University.[6] He had two older sisters, Marianna and Sarah. Marianna became a biochemist, and was a significant role model for Fred, who delighted in the smells and explosions produced by chemistry sets in that era.[7] He attended high school at Phillips Exeter Academy, and later recalled that "the excellent science department even permitted certain students the unsupervised run of the laboratories outside of class hours. This attitude played a strong role... in cementing our commitment to scientific careers."[6] He learned glassblowing and electronics there, and tried to measure the universal gravitational constant using 100-pound cannonballs.[6]
With strong science interests, Richards thwarted his family's expectations by choosing MIT (Massachusetts Institute of Technology) rather than Yale for college in 1943, majoring in chemistry. His undergraduate time was interrupted by two years in the army, which he described as "uneventful".[6] He then joined the Biochemistry department at Harvard Medical School and the lab of Barbara Low. She had worked with Dorothy Crowfoot Hodgkin to solve the x-ray crystal structure of penicillin, and was later active in protein crystallography. The phase problem had not yet been solved to allow determination of protein structure, so his Ph.D. thesis (completed in 1952) studied the density and solvent content in crystals to help determine very accurate molecular weights for proteins. In 1954 he went to the Carlsberg Laboratory in Copenhagen to do postdoctoral research with Kaj Linderstrøm-Lang, where he started his classic work on the ribonuclease enzyme.[7] He also absorbed the scientific and mentorship style of Lang, who Richards called "a delightful individual, full of fun and jokes as well as science" exemplifying "simple, inexpensive, ingenious, and insightful experiments".[8] In 1955 Richards joined the faculty at Yale University, where he stayed for the rest of his career.

Richards was an avid and enthusiastic sailor. In addition to sailing on Long Island Sound, he voyaged north along the Canadian coast, south to Bermuda, and even across the Atlantic several times with a small crew of family and friends.[4][9] He and his wife had sailboats (Hekla 1 and 2) and an outboard-motor utility boat known as "Sally's Baage"[10] (the spelling presumably a comment on her Maine accent), which he had built himself.[9] Chris Anfinsen, Richards's friend and his colleague as editors of Advances in Protein Chemistry[9] and who recommended the Carlsberg Lab to him,[6] was also an avid sailor, and they sometimes joined forces.[9] Wendell Lim wrote that, "a dedicated sailor since childhood, Fred almost always took a month off each summer to captain a major sailing excursion, returning to lab afterwards refreshed and ready to work. His sailing adventures included several transatlantic voyages. He was also an avid ice hockey player."[7]
Richards lived in Guilford, Connecticut, a coastal town about 10 miles east of New Haven, situated between the Metacomet Ridge and Long Island Sound. Fred was married twice, to Heidi Clark Richards, daughter of biochemist Hans Clarke,[11][12] and in 1959 to Sarah (Sally) Wheatland Richards, a marine biologist.[10] He had three children – Sarah, Ruth, and George – and four grandchildren.[10] His daughter Sarah described him as "a lifelong scientist and sailor.... His main loves were his scientific work which he finished at Yale University, sailing, working in his shop, and helping in the community."[11] Fred and Sally were a major presence in local land conservation efforts, both on committees and in working projects out on the land and water.[10][13] He donated a 41-acre shoreline property to the Yale Peabody Museum Natural Areas, which they described as "one of the few natural forest areas left in the state." The property now has long-term protection for use in biological and geological research.[14]
Research career
Two-component ribonuclease S system
On December 2, 1957, at Yale University, Richards performed a simple experiment on the protein Ribonuclease A (RNase A) that helped change the scientific community's view of the physical nature of protein molecules.[6] Using a particular protease (Subtilisin), RNase A was converted into a split protein (RNase S), which is composed of two parts called S-peptide and S-protein (Richards & Vithayathil 1959). Richards had developed that cleavage system as a postdoc at the Carlsberg Laboratory in Sweden, using purified ribonuclease protein that had been donated to Christian Anfinsen by the Armour Company and that Anfinsen shared with Richards and other researchers.[4][8] Richards found that, when separated, S-protein and S-peptide had no RNase activity, but that the RNase enzymatic activity was restored when the parts were recombined in the test tube.[15] In an autobiographical piece, Richards wrote that "this discovery came as a surprise to the scientific community at that time.... In retrospect, this may have been the high point of my career in terms of excitement."[6] This experiment showed that proteins maintain 3-dimensional order and tight binding between their interacting parts and that the structural information is inherent in the protein itself, foreshadowing both Anfinsen's later work showing that sequence determines structure[16] and also the idea that hormones or other small molecules can bind tightly and specifically to proteins,[6] a concept basic to how pharmaceutical companies design drugs today. Two years later, the protein structure of myoglobin confirmed such specific 3D relationships.[17] Later, with Marilyn Doscher and Flo Quiocho, Richards demonstrated that ribonuclease S as well as carboxypeptidase were enzymatically active in the crystals, important evidence to silence doubts that the conformations of proteins in crystals are directly relevant to their biological activity in cells.[7][18][19]
Ribonuclease crystal structure
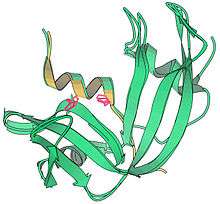
Along with colleague Harold W. Wyckoff, who had worked on early research toward the myoglobin structure,[20] the effort to solve the RNase S 3-dimensional structure was spearheaded by Richards. Done in 1966 and published in 1967, the analyses of RNase S [21] and RNase A[22] jointly made ribonuclease the third distinct protein structure to be determined by X-ray diffraction of crystals, after myoglobin/hemoglobin and hen-egg lysozyme,[23] and the first to be done in the United States. Later, the Yale group collected more diffraction data, and in 1970 published the RNase S structure in full detail at 2.0 Å resolution (Wyckoff et al., 1970). Coordinates for ribonuclease S were deposited into the international Protein Data Bank in 1973 as PDB: 1RNS, among the first small set of macromolecular structures.[24]
The black-and-white ribbon drawing above shows the large, twisted beta sheet (arrows) of ribonuclease, flanked by several alpha-helices (spirals). The shorter S-peptide piece is behind, starting at upper left with a helix and ending with the chain break (between residues 20-21) at lower right.[5] The active site for RNA cleavage (in the groove at center front in this drawing) involves one histidine side chain from the S-peptide fragment and another from the S-protein part.[21] The computer image shows superimposed structures of ribonuclease S and A, with the S-peptide in gold and the active site histidines in hot pink. The close match of the 3D structures shows that the 2-fragment S system does indeed fold to the active form (Wyckoff et al., 1970).
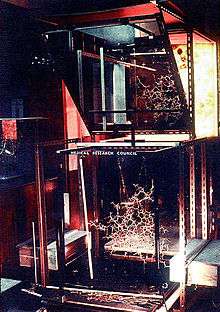
"Richards' box"
In 1968, while on sabbatical with David Phillips at Oxford, Richards developed a large optical comparator device called a "Richards' box" (or "Fred's Folly") which enabled crystallographers to build physical models of protein structures by viewing the stacked sheets of electron density through a half-silvered mirror (see photo).[25] Once the Folly had been constructed, he built an all-atom brass model of RNase S quite rapidly.[6] This was the method of choice for building protein crystallographic models into electron density until the late 1970s, when it was superseded by molecular computer graphics programs such as Grip-75[26] and then Frodo.[27]
Richards showed his sense of humor in a later review of developments in the use and construction of Richards boxes.[28] He provided a "correction to the Original Bibliographic Citations," complete with diagrams, for a theatrical stage technique that used selective illumination and a sheet of plate glass inclined at 45° to give an illusion of the nymph Amphitrite rising from the sea and floating in air, or of an audience volunteer dissolving to a skeleton and back again. Richards ended that section by noting that "had this reference been known to the author in 1968 no further description of the 'folly' would have been required."
Solvent-accessible surface and molecular packing
Richards' most enduring long-term scientific interest was in protein folding and packing, studied both experimentally and theoretically, and mostly from a geometrical perspective. As summarized by George Rose, "protein folders can be divided into 'minimizers' and 'packers'. The former seek to minimize the interaction energy among atoms or groups of atoms, whereas the latter concentrate on probable geometry, guided by both excluded volume limitations and structural motifs seen in proteins of known structure." Fred was a founding influence for the packers, who built on his observations about packing density, areas, and volumes.[9]
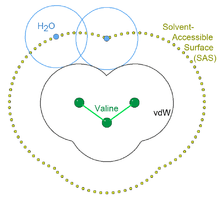
In 1971, with Byungkook Lee, Richards introduced the concept and a quantitative measure for the solvent-accessible surface (SAS) of amino acid residues in folded protein structures (Lee & Richards 1971). The surface is constructed by tracing the center of an imaginary ball, its radius that of a water molecule (taken as 1.4 Å), as it rolls over the van der Waals surfaces of the proteins. Thus defined, the surface is continuous and each point on it is unambiguously associated with a specific protein atom (the nearest). The Lee & Richards definition has been widely adopted as the standard measure for solvent accessibility, for instance to evaluate exposure per residue as a percentage of accessible vs total surface area,[29] and as the basis of the buried-surface-area method for estimating the energetics of protein/protein contacts.[30]
Richards 1974 introduced the Voronoi polyhedra construction to protein chemistry, a contribution reviewed more recently by Gerstein and Richards.[31] This approach has been adopted by many others[32][33] and has been put on a firm mathematical footing by the work of Herbert Edelsbrunner.[34] With Jay Ponder in 1987, as part of an exploration of using internal packing of sidechains to enumerate the possible sequences compatible with a given protein backbone structure (a foreshadowing of protein engineering and design), Richards developed the first side-chain rotamer library. (Ponder & Richards 1987) Increasingly detailed rotamer libraries have been made since then by other research groups, with some used primarily for structure validation[35] and others for homology modeling or protein design.[36] With Craig Kundrot, Richards investigated the effects of high pressure (1000 atmospheres) on protein structure, using hen-egg lysozyme crystals,[37] finding that the structure was robust to such pressures apart from a quite modest compaction in size. In the 1990s, Richards and collaborators used a combination of theory and experiment to investigate how the well-packed interior of proteins can nevertheless accommodate mutations.[38][39][40]
Other research areas
In the 1970s, with a succession of students and postdocs, the lab developed a series of chemical, photochemical, and cross-link labels for determining the position and relationships of proteins in biological membranes (Peters & Richards 1977),[41] including glutaraldehyde[42] and what was one of the two first general uses of the exceptionally tight interaction of biotin with avidin,[6] anchored to ferritin for use in electron microscopy.[43][44][45] The biotin–avidin system quickly became a central method in cell biology, immunology, and protein engineering, as well as electron microscopy.[46]
With David Wishart and Brian Sykes, he developed the chemical shift index for NMR assignment of protein secondary structure (Wishart, Sykes & Richards 1991 & Wishart, Sykes & Richards 1992). This is still considered a standard tool in the NMR field.[47] Separately, around 1990, Homme Hellinga, with Richards, developed computational tools to design metal-binding sites into proteins,[48] and used them to build a new metal site into thioredoxin.[49]
Richards is named as a depositor on 27 crystal structure entries in the Protein Data Bank, including the now-obsoleted ribonuclease S (PDB: 1RNS), hen egg lysozyme (PDB: 2LYM), SH3 domains (PDB: 1SEM), the ion-channel-forming alamethicin (PDB: 1AMT) (Fox & Richards 1982), and mutants of ribonuclease S (e.g., PDB: 1RBD;PDB: 1RBI), of Staphylococcal nuclease (e.g., PDB: 1NUC;PDB: 1A2T), and of lambda repressor in complex with DNA (PDB: 1LLI).
Administration, mentoring, and outside activities
The Department of Molecular Biophysics and Biochemistry ("MB&B")[50] that Richards founded and chaired at Yale, which amalgamated the medical school Biochemistry and the university Molecular Biophysics departments, was considered to have "quickly gained pre-eminent stature."[9] Many of those faculty became members of the National Academy of Sciences,[4][9] and Tom Steitz shared the Nobel Prize in 2009 for crystal structures of the ribosome.[51] Richards was known as a highly valued mentor and friend to students, faculty, and colleagues, including a very supportive approach to women and African–Americans, according to Jim Staros.[12] His colleague George D. Rose wrote that Richards' lectures were insightful, delivered with clarity and humor, and often deliberately provocative, and that Richards worked to improve the scientific community in general.[9] For instance, in the late 1980s, he was the primary author, and the first of many signers, of a widely circulated letter that successfully urged a policy of depositing 3D atomic coordinates on scientific journals, on the NIH, and on individual crystallographers.[6][52] He also lobbied, less successfully, for a let-up in overall publication pressure but an increased emphasis on a few first-class papers, by having promotion committees only consider a list of 12 key papers.[6]
Summary of career events
- 1954, NRC Postdoctoral Fellow, Carlsberg Laboratory, Denmark[8]
- 1955, joined Yale faculty, in Biochemistry at the Medical School[12]
- 1963, Professor and chair of the Department of Molecular Biophysics at Yale University[6]
- 1965, Pfizer – Paul-Lewis Award in Enzyme Chemistry
- 1968, Member, American Academy of Arts and Sciences[53]
- 1969–73, founding chair of the Department of Molecular Biophysics and Biochemistry at Yale[6]
- 1971, Member, National Academy of Sciences
- 1972, President of the Biophysical Society[54]
- 1976–91, Director of the Jane Coffin Childs Fund for Medical Research[6]
- 1978, Kaj Linderstrøm-Lang Prize in Protein Chemistry
- 1979, President of the ASBMB[12]
- 1988, American Society for Biochemistry and Molecular Biology – Merck Award [55]
- 1988, Protein Society – Stein and Moore Award [56]
- 1995, Connecticut Medal of Science[4]
Highly cited papers
Articles with over 500 citations according to Web of Science as of June 18, 2012:[57]
- Lee, B.; Richards, F.M. (1971). "The interpretation of protein structures: Estimation of static accessibility". Journal of Molecular Biology. 55 (3): 379–400. doi:10.1016/0022-2836(71)90324-X. PMID 5551392.
- Richards, F.M. (1977). "Areas, Volumes, Packing, and Protein Structure". Annual Review of Biophysics and Bioengineering. 6: 151–176. doi:10.1146/annurev.bb.06.060177.001055. PMID 326146.
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. (1992). "The chemical shift index: A fast and simple method for the assignment of protein secondary structure through NMR spectroscopy". Biochemistry. 31 (6): 1647–1651. doi:10.1021/bi00121a010. PMID 1737021.
- Wishart, D.S.; Sykes, B.D.; Richards, F.M. (1991). "Relationship between nuclear magnetic resonance chemical shift and protein secondary structure". Journal of Molecular Biology. 222 (2): 311–333. doi:10.1016/0022-2836(91)90214-Q. PMID 1960729.
- Ponder, J.W.; Richards, F.M. (1987). "Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes". Journal of Molecular Biology. 193 (4): 775–791. doi:10.1016/0022-2836(87)90358-5. PMID 2441069.
- Richards, F.M. (1974). "The interpretation of protein structures: Total volume, group volume distributions and packing density". Journal of Molecular Biology. 82 (1): 1–14. doi:10.1016/0022-2836(74)90570-1. PMID 4818482.
- Richards, F.M.; Vithayathil, P.J. (1959). "The preparation of subtilisin-modified ribonuclease and the separation of the peptide and protein components". Journal of Biological Chemistry. 234 (6): 1459–1465. PMID 13654398.
- Fox, R.O.; Richards, F.M. (1982). "A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-Å resolution". Nature. 300 (5890): 325–330. Bibcode:1982Natur.300..325F. doi:10.1038/300325a0. PMID 6292726.
- Peters, K.; Richards, F. M. (1977). "Chemical Cross-Linking: Reagents and Problems in Studies of Membrane Structure". Annual Review of Biochemistry. 46: 523–551. doi:10.1146/annurev.bi.46.070177.002515. PMID 409338.
- Wyckoff, H.W.; Tsernoglou, D.; Hanson, A.W.; Knox, J.R.; Lee, B.; Richards, F.M. (1970). "The three-dimensional structure of ribonuclease-S. Interpretation of an electron density map at a nominal resolution of 2 Å". Journal of Biological Chemistry. 245 (2): 305–328. PMID 5460889.
References
- ↑
- ↑ Fred Richard's Ph.D. Thesis. OCLC 76995296.
- 1 2 3 4
- 1 2 3 4 5 Steitz, TA (2009). "Retrospective. Frederic M. Richards (1925–2009)". Science. 323 (5918): 1181. doi:10.1126/science.1171157. PMID 19251620.
- 1 2 Richardson, JS (1981). "The Anatomy and Taxonomy of Proteins". Advances in Protein Chemistry. 34: 167–339 . doi:10.1016/S0065-3233(08)60520-3. PMID 7020376.
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 Richards FM (1997). "Whatever Happened to the Fun? An Autobiographical Investigation". Annual Review of Biophysics and Biomolecular Structure. 26: 1–25. doi:10.1146/annurev.biophys.26.1.1.
- 1 2 3 4 Lim WA (2009). "Frederic M Richards 1925–2009". Nature Structural & Molecular Biology. 16 (3): 230–232. doi:10.1038/nsmb0309-230. PMID 19259121.
- 1 2 3 Richards FM (1992). "Linderstrøm-Lang and the Carlsberg Laboratory: the view of a postdoctoral fellow in 1954". Protein Science. 1: 1721–1730. doi:10.1002/pro.5560011221. PMC 2142135
 . PMID 1304902.
. PMID 1304902. - 1 2 3 4 5 6 7 8 Rose GD (2009). "In Memoriam: Frederic M. Richards (1925–2009)". Proteins. 75 (3): 535–539. doi:10.1002/prot.22400. PMID 19226638.
- 1 2 3 4 "Sally Richards Remembered". Guilford Land Conservation Trust. Retrieved July 9, 2012.
- 1 2 "Frederic M. Richards". Retrieved 2009-01-14.
- 1 2 3 4 Staros JV. "Retrospective: Frederic M. Richards (1925–2009)" (PDF). ASBMB Today. April 2009: 16–18.
- ↑ "An Incomparable Couple". Guilford Land Conservation Trust. Retrieved July 9, 2012.
- ↑ "Yale Peabody Museum Natural areas: Richards property". Yale Peabody Museum. Retrieved July 10, 2012.
- ↑ Richards FM (1958). "On the enzymic activity of subtilisin-modified ribonuclease". Proceedings of the National Academy of Sciences of the United States of America. 44: 162–166. Bibcode:1958PNAS...44..162R. doi:10.1073/pnas.44.2.162. PMC 335382
 . PMID 16590160.
. PMID 16590160. - ↑ Fox RO (February 2009). "Obituary: Frederic Richards (1925–2009)". Nature. 457 (7232): 976. Bibcode:2009Natur.457..976F. doi:10.1038/457976a. PMID 19225517.
- ↑ Kendrew JC, Dickerson RE, Strandberg BE, Hart RG, Davies DR, Phillips DC, Shore VC (1960). "Structure of myoglobin: a three-dimensional Fourier synthesis at 2 Å. resolution". Nature. 185 (4711): 422–427. Bibcode:1960Natur.185..422K. doi:10.1038/185422a0. PMID 18990802.
- ↑ Doscher MS, Richards FM (1963). "The activity of an enzyme in the crystalline state: Ribonuclease-S". Journal of Biological Chemistry. 238: 2399–2406.
- ↑ Quiocho FA, Richards FM (1964). "Intermolecular cross linking of a protein in the crystalline state: carboxypeptidase-a". Proceedings of the National Academy of Sciences of the United States of America. 52 (3): 833–839. Bibcode:1964PNAS...52..833Q. doi:10.1073/pnas.52.3.833. PMC 300354
 . PMID 14212562.
. PMID 14212562. - ↑ Bodo G, Dintzis HM, Kendrew JC, Wyckoff HW (1959). "The crystal structure of myoglobin. V. A low resolution three-dimensional Fourier synthesis of sperm-whale myoglobin crystals". Proceedings of the Royal Society A. 253 (1272): 70–102. Bibcode:1959RSPSA.253...70B. doi:10.1098/rspa.1959.0179.
- 1 2 Wyckoff HW, Hardman KD, Allewell NM, Inagami T, Johnson LN, Richards FM (1967). "The structure of ribonuclease-S at 3.5 Å resolution". Journal of Biological Chemistry. 242: 3984–3988. PMID 6037556.
- ↑ Kartha G, Bello J, Harker D (1967). "Tertiary Structure of Ribonuclease". Nature. 213 (5079): 862–865. Bibcode:1967Natur.213..862K. doi:10.1038/213862a0. PMID 6043657.
- ↑ Blake CCF, Koenig DF, Mair GA, North ACT, Phillips DC, Sarma VR (1965). "Structure of hen egg-white lysozyme: a three-dimensional Fourier synthesis at 2 Å resolution". Nature. 206 (4986): 757–761. Bibcode:1965Natur.206..757B. doi:10.1038/206757a0. PMID 5891407.
- ↑ Bernstein FC, Koetzle T F, Williams, GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977). "The protein data bank: A computer-based archival file for macromolecular structures". Journal of Molecular Biology. 112: 535–542. doi:10.1016/S0022-2836(77)80200-3.
- ↑ Richards FM (1968). "The matching of physical models to three-dimensional electron-density maps: a simple optical device". Journal of Molecular Biology. 37 (1): 225–230. doi:10.1016/0022-2836(68)90085-5. PMID 5760491.
- ↑ Brooks FP Jr (1977). "The Computer "Scientist" as Toolsmith: Studies in Interactive Computer Graphics". Proceedings of IFIP (International Federation for Information Processing) Congress. North Holland: 625–634.
- ↑ Jones TA (1978). "A Graphics Model Building and Refinement System for Macromolecules". Journal of Applied Crystallography. 11: 268–272. doi:10.1107/S0021889878013308.
- ↑ Richards FM (1985). "Optical Matching of Physical Models and Electron Density Maps: Early Developments". Methods in Enzymology. 115: 145–154. doi:10.1016/0076-6879(85)15012-3. PMID 3908881.
- ↑ Shrake A, Rupley JA (1973). "Environment and exposure to solvent of protein atoms. Lysozyme and insulin". Journal of Molecular Biology. 79 (2): 351–71. doi:10.1016/0022-2836(73)90011-9. PMID 4760134.
- ↑ Chothia C (1976). "The nature of the accessible and buried surfaces in proteins". Journal of Molecular Biology. 105 (1): 1–12. doi:10.1016/0022-2836(76)90191-1. PMID 994183.
- ↑ M Gerstein; F M Richards (2001). "Protein Geometry: volumes, areas, and distances.". International Tables for Crystallography. F: 531–539.
- ↑ Gerstein M, Tsai J, Levitt M (1995). "The volume of atoms on the protein surface: calculated from simulation, using Voronoi polyhedra". Journal of Molecular Biology. 249 (5): 955–966. doi:10.1006/jmbi.1995.0351. PMID 7540695.
- ↑ Poupon A (2004). "Voronoi and Voronoi-related tessellations in studies of protein structure and interaction". Current Opinion in Structural Biology. 14: 233–41. doi:10.1016/j.sbi.2004.03.010.
- ↑ Edelsbrunner H, Mucke EP (1994). "Three-dimensional alpha shapes". ACM Transactions on Graphics. 13: 43–72. doi:10.1145/174462.156635.
- ↑ Read RJ, Adams PD, Arendall WB III, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lütteke T, Otwinowski Z, Perrakis A, Richardson JS, Sheffler WH, Smith JL, Tickle IJ, Vriend G, Zwart PH (2011). "A New Generation of Crystallographic Validation Tools for the Protein Data Bank". Structure. 19: 1395–1412. doi:10.1016/j.str.2011.08.006. PMC 3195755
 . PMID 22000512.
. PMID 22000512. - ↑ Dunbrack RL Jr. (2002). "Rotamer libraries in the 21st century". Current Opinion in Structural Biology. 12 (4): 431–40. doi:10.1016/S0959-440X(02)00344-5. PMID 12163064.
- ↑ Kundrot CE, Richards FM (1987). "Crystal structure of hen egg-white lysozyme at a hydrostatic pressure of 1000 atmospheres". Journal of Molecular Biology. 193: 157–170. doi:10.1016/0022-2836(87)90634-6.
- ↑ Varadarajan R, Richards, FM (1992). "Crystallographic structures of ribonuclease S variants with nonpolar substitution at position 13: packing and cavities". Biochemistry. 31 (49): 12315–12327. doi:10.1021/bi00164a005. PMID 1463720.
- ↑ Lim WA, Hodel A, Sauer RT, Richards FM (1994). "The crystal structure of a mutant protein with altered but improved hydrophobic core packing". Proceedings of the National Academy of Sciences of the United States of America. 91 (1): 423–427. Bibcode:1994PNAS...91..423L. doi:10.1073/pnas.91.1.423. PMC 42960
 . PMID 8278404.
. PMID 8278404. - ↑ Wynn R, Harkins PC, Richards FM, Fox RO (1996). "Mobile unnatural amino acid side chains in the core of staphylococcal nuclease". Protein Science. 5 (6): 1026–1031. doi:10.1002/pro.5560050605. PMC 2143447
 . PMID 8762134.
. PMID 8762134. - ↑ Staros JV, Richards FM (1974). "Photochemical labeling of the surface proteins of human erythrocytes". Biochemistry. 13 (13): 2720–2726. doi:10.1021/bi00710a010. PMID 4847541.
- ↑ Richards FM, Knowles JR (1968). "Glutaraldehyde as a protein cross-linking reagent". Journal of Molecular Biology. 37: 231–233. doi:10.1016/0022-2836(68)90086-7.
- ↑ Heitzmann H, Richards FM (1974). "Use of the avidin–biotin complex for specific staining of biological membranes in electron microscopy". Proceedings of the National Academy of Sciences of the United States of America. 71 (9): 3537–3541. Bibcode:1974PNAS...71.3537H. doi:10.1073/pnas.71.9.3537. PMC 433809
 . PMID 4139715.
. PMID 4139715. - ↑ Manning, J. E.; Hershey, N. D.; Broker, T. R.; Pellegrini, M.; Mitchell, H. K.; Davidson, N. (1975). "A new method of in situ hybridization". Chromosoma. 53 (2): 107–117. doi:10.1007/bf00333039. PMID 172297.
- ↑ Richards FM (1990). "Reflections". Methods in Enzymology. 184: 3–5. doi:10.1016/0076-6879(90)84255-F. PMID 2201877.
- ↑ Diamandis EP, Christopoulos TK (1991). "The biotin-(strept)avidin system: principles and applications in biotechnology". Clinical Chemistry. 37 (5): 625–636. PMID 2032315.
- ↑ Cavanagh, John; Fairbrother, Wayne J.; Palmer, Arthur G. III; Skelton, Nicholas J. (2006). Protein NMR Spectroscopy: Principles and Practice (2nd ed.). Academic Press. ISBN 0-12-164491-X.
- ↑ Hellinga HW, Richards FM (1991). "Construction of new ligand-binding sites in proteins of known structure.1. Computer-aided modeling of sites with predefined geometry". Journal of Molecular Biology. 222: 763–785. doi:10.1016/0022-2836(91)90510-D.
- ↑ Hellinga HW, Carradona JP, Richards FM (1991). "Construction of new ligand-binding sites in proteins of known structure. 2. Grafting of a buried transition-metal binding site into E coli thioredoxin". Journal of Molecular Biology. 222: 787–803. doi:10.1016/0022-2836(91)90511-4.
- ↑ http://www.mbb.yale.edu/
- ↑ "The Nobel Prize in Chemistry 2009". Nobelprize.org. Retrieved 21 May 2012.
- ↑ Richards FM (1988). "Public access to x-ray diffraction data". Journal of Computer-Aided Molecular Design. 2 (1): 3–4. Bibcode:1988JCAMD...2....3R. doi:10.1007/BF01532048. PMID 3199147.
- ↑ "R". Members of the American Academy of Arts & Sciences: 1780–2011 (PDF). American Academy of Arts & Sciences. 2012. p. 500.
- ↑ "Past Officers". Biophysical Society. Retrieved 19 June 2012.
- ↑ "ASBMB-Merck Award". ASBMB. Retrieved 18 June 2012.
- ↑ "Past award recipients". Protein Society. Retrieved 18 June 2012.
- ↑ "Web of Science". Thomson Reuters. Retrieved 18 June 2012.
Further reading
- Ben Lillie, "What Wikipedia Taught Me About My Grandfather," The Atlantic, Nov. 18, 2014.
Podcast
- Ben Lillie, "The Truth About My Grandfather," "The Story Collider," August 26, 2016.
External links
- Richards biography on Proteopedia, by Eric Martz
- Department of Molecular Biophysics & Biochemistry, Yale University