Candida albicans
| Candida albicans | |
|---|---|
 | |
| Scientific classification | |
| Kingdom: | Fungi |
| Division: | Ascomycota |
| Class: | Saccharomycetes |
| Order: | Saccharomycetales |
| Family: | Saccharomycetaceae |
| Genus: | Candida |
| Species: | C. albicans |
| Binomial name | |
| Candida albicans (C.P.Robin) Berkhout (1923) | |
| Synonyms | |
Candida albicans is a dimorphic fungus that grows both as yeast and filamentous cells and one of the few species of the Candida genus that cause the infection candidiasis in humans.[3][4] C. albicans is responsible for 50–90% of all cases of candidiasis in humans.[4] Systemic fungal infections (fungemias) including those by C. albicans have emerged as important causes of morbidity and mortality in immunocompromised patients (e.g., AIDS, cancer chemotherapy, organ or bone marrow transplantation). C. albicans biofilms may form on the surface of implantable medical devices. In addition, hospital-acquired infections by C. albicans have become a cause of major health concerns. About 85-95% of vaginal infections cases are responsible for physician office visits every year.[5]
C. albicans is a common member of human gut flora and is detectable in the gastrointestinal tract in 40% of healthy adults.[3][4][6] It is usually a commensal organism, but can become pathogenic in immunocompetent individuals under a variety of conditions.[3][4] Overgrowth of the fungus results in candidiasis (candidosis).[3][4] Candidiasis is often observed in immunocompromised individuals, including HIV-infected patients. It commonly occurs on mucous membranes in the mouth or vagina, but may affect a number of other regions. For example, higher prevalence of colonization of C. albicans was reported in young individuals with tongue piercing, in comparison to unpierced matched individuals.[7] To infect host tissue, the usual unicellular yeast-like form of C. albicans reacts to environmental cues and switches into an invasive, multicellular filamentous form, a phenomenon called dimorphism.[8] In addition, an overgrowth infection is considered superinfection, usually applied when an infection become opportunistic and very resistant to antifungals. It then becomes suppressed by antibiotics. The infection is prolonged when the original sensitive strain is replaced by the antibiotic-resistant strain.[9]
Genome
One of the most important features of the C. albicans genome is the occurrence of numeric and structural chromosomal rearrangements as means of generating genetic diversity, named chromosome length polymorphisms (contraction/expansion of repeats), reciprocal translocations, chromosome deletions and trisomy of individual chromosomes. These karyotypic alterations lead to changes in the phenotype, which is an adaptation strategy of this fungus. These mechanisms will be better understood with the complete analysis of the C. albicans genome.
An unusual feature of the Candida genus is that in many of its species (including C. albicans and C. tropicalis, but not, for instance, C. glabrata) the CUG codon, which normally specifies leucine, specifies serine in these species. This is an unusual example of a departure from the standard genetic code, and most such departures are in start codons or, for eukaryotes, mitochondrial genetic codes.[10][11][12] This alteration may, in some environments, help these Candida species by inducing a permanent stress response, a more generalized form of the heat shock response.[13]
The genome of C. albicans is highly dynamic, and this variability has been used advantageously for molecular epidemiological studies and population studies in this species. The genome sequence has allowed for identifying the presence of a parasexual cycle (no detected meiotic division) in C. albicans.[14] This study of the evolution of sexual reproduction in six Candida species found recent losses in components of the major meiotic crossover-formation pathway, but retention of a minor pathway.[14] The authors suggested that if Candida species undergo meiosis it is with reduced machinery, or different machinery, and indicated that unrecognized meiotic cycles may exist in many species. In another evolutionary study, introduction of partial CUG identity redefinition (from Candida species) into Saccharomyces cerevisiae clones caused a stress response that negatively affected sexual reproduction. This CUG identity redefinition, occurring in ancestors of Candida species, was thought to lock these species into a diploid or polyploid state with possible blockage of sexual reproduction.[15]
Dimorphism
Although often referred to as "dimorphic", C. albicans is in fact polyphenic. When cultured in standard yeast laboratory medium, C. albicans grows as ovoid "yeast" cells. However, mild environmental changes in temperature and pH can result in a morphological shift to pseudohyphal growth.[16] Pseudohyphae share many similarities with yeast cells,[17] but their role during candidiasis remains unknown. When C. albicans cells are grown in a medium that mimics the physiological environment of a human host, they grow as "true" hyphae. Its ability to form hyphae has been proposed as a virulence factor, as these structures are often observed invading tissue, and strains that are unable to form hyphae are defective in causing infection. Candida albicans can also form Chlamydospores, the function of which remains unknown.[18]
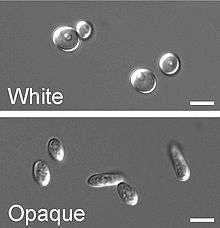
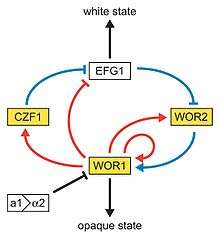
In a process that superficially resembles dimorphism, C. albicans undergoes a process called phenotypic switching, in which different cellular morphologies are generated spontaneously. Of the classically studied strains, one that undergoes phenotypic switching is WO-1,[19] which consists of two phases: one that grows as round cells in smooth, white colonies and one that is rod-like and grows as flat, gray colonies. The other strain known to undergo switching is 3153A; this strain produces at least seven different colony morphologies. In both the WO-1 and 3153A strains, the different phases convert spontaneously to the other(s) at a low frequency. The switching is reversible, and colony type can be inherited from one generation to another. While several genes that are expressed differently in different colony morphologies have been identified, some recent efforts focus on what might control these changes. Further, whether a potential molecular link between dimorphism and phenotypic switching occurs is a tantalizing question.[20]
In the 3153A strain, a gene called SIR2 (for silent information regulator), which seems to be important for phenotypic switching, has been found. SIR2 was originally found in Saccharomyces cerevisiae (brewer's yeast), where it is involved in chromosomal silencing—a form of transcriptional regulation, in which regions of the genome are reversibly inactivated by changes in chromatin structure (chromatin is the complex of DNA and proteins that make chromosomes). In yeast, genes involved in the control of mating type are found in these silent regions, and SIR2 represses their expression by maintaining a silent-competent chromatin structure in this region. The discovery of a C. albicans SIR2 implicated in phenotypic switching suggests it, too, has silent regions controlled by SIR2, in which the phenotype-specific genes may reside.
Another potential regulatory molecule is Efg1p, a transcription factor found in the WO-1 strain that regulates dimorphism, and more recently has been suggested to help regulate phenotypic switching. Efg1p is expressed only in the white and not in the gray cell-type, and overexpression of Efg1p in the gray form causes a rapid conversion to the white form.[21][22]
So far, very few data suggest dimorphism and phenotypic switching use common molecular components. However, it is not inconceivable that phenotypic switching may occur in response to some change in the environment, as well as being a spontaneous event. How SIR2 itself is regulated in S. cerevisiae may yet provide clues as to the switching mechanisms of C. albicans.
Heterozygosity
The heterozygosity of the Candida genome exceeds that found in other genomes and is widespread among clinical isolates. Nonsynonymous single-base polymorphisms result in two proteins that differ in one or several amino acids that may confer functional differences for each protein. This situation considerably increases the number of different proteins encoded by the genome.[23]
Proteins important for pathogenesis
Hwp1
Hwp1 stands for Hyphal wall protein 1. Hwp1 is a mannoprotein located on the surface of the hyphae in the hyphal form of Candida albicans. Hwp1 is a mammalian transglutaminase substrate. This host enzyme allows Candida albicans to attach stably to host epithelial cells.[24] Adhesion of Candida albicans to host cells is an essential first step in the infection process for colonization and subsequent induction of mucosal infection.
Slr1
RNA-binding protein Slr1 was recently discovered to play a role in instigating the hyphal formation and virulence in C. albicans.[25]
Candidalysin
Candidalysin is a cytolytic 31-amino acid α-helical peptide toxin that is released during hyphal formation. It contributes to virulence during mucosal infections.[26]
Epidemiology
Candida is found worldwide but most commonly compromises immunocompromised individuals diagnosed with serious diseases such as HIV and cancer. Candida are ranked as one of the most common groups of organisms that cause noscomial infections. Especially high risk individuals are patients in Intensive Care Units (ICU), have recently undergone surgery or transplant.[27] Candida albicans infections is the top source of fungal infections in critically ill or otherwise immuncompromised patients.[28] These patients predominantly develop oropharyngeal or thrush candidiasis, which can lead to malnutrition and interfere with the absorption of medication.[29] Parts of the body that are commonly infected include the skin, genitals, throat, mouth, and blood. Candida continues to be the fourth most commonly isolated organism in bloodstream infections (BSIs).[30] Furthermore, Candida albicans bloodstream infections have been associated with a high mortality rate.[31] Methods of transmission include mother to infant through childbirth, people-to-people acquired infections that most commonly occur in hospital settings where immunocompromised patients acquire the yeast from healthcare workers and has a 40% incident rate. Men can become infected after having sex with a woman that has an existing vaginal yeast infection.[27] Although Candida albicans is the most common cause of candidemia, there has been a decrease in the incidence and an increases isolation of non-albicans species of Candida in recent years.[32] Preventive measures include keeping a healthy lifestyle including good nutrition, proper nutrition, and careful antibiotic use.
Application in engineering
Candida albicans has been used in combination with carbon nanotubes (CNT) to produce stable electrically conductive bio-nano-composite tissue materials that have been used as temperature sensing elements[33]
Treatment
Treatment commonly includes:[34]
- amphotericin B, echinocandin, or fluconazole for systemic infections
- Nystatin for oral and esophageal infections
- Clotrimazole for skin and genital yeast infections[35]
See also
- Intestinal permeability
- Torula yeast (Candida utilis)
- Neonatal infection
References
- ↑ Candida albicans at NCBI Taxonomy browser, url accessed 2006-12-26
- ↑ "McClary, Dan Otho (May 1952). "Factors Affecting the Morphology of Candida Albicans". Annals of the Missouri Botanical Garden. 39 (2): 137–164. doi:10.2307/2394509. JSTOR 2394509.
- 1 2 3 4 Erdogan A, Rao SS (April 2015). "Small intestinal fungal overgrowth". Curr Gastroenterol Rep. 17 (4): 16. doi:10.1007/s11894-015-0436-2. PMID 25786900.
Small intestinal fungal overgrowth (SIFO) is characterized by the presence of excessive number of fungal organisms in the small intestine associated with gastrointestinal (GI) symptoms. Candidiasis is known to cause GI symptoms particularly in immunocompromised patients or those receiving steroids or antibiotics. However, only recently, there is emerging literature that an overgrowth of fungus in the small intestine of non-immunocompromised subjects may cause unexplained GI symptoms. Two recent studies showed that 26 % (24/94) and 25.3 % (38/150) of a series of patients with unexplained GI symptoms had SIFO. The most common symptoms observed in these patients were belching, bloating, indigestion, nausea, diarrhea, and gas. The underlying mechanism(s) that predisposes to SIFO is unclear but small intestinal dysmotility and use of proton pump inhibitors has been implicated. However, further studies are needed; both to confirm these observations and to examine the clinical relevance of fungal overgrowth, both in healthy subjects and in patients with otherwise unexplained GI symptoms. ... For routine SIFO in an immunocompetent host, a 2–3 week oral course of fluconazole 100–200 mg will suffice.
- 1 2 3 4 5 Martins N, Ferreira IC, Barros L, Silva S, Henriques M (June 2014). "Candidiasis: predisposing factors, prevention, diagnosis and alternative treatment". Mycopathologia. 177 (5-6): 223–240. doi:10.1007/s11046-014-9749-1. PMID 24789109.
Candida species and other microorganisms are involved in this complicated fungal infection, but Candida albicans continues to be the most prevalent. In the past two decades, it has been observed an abnormal overgrowth in the gastrointestinal, urinary and respiratory tracts, not only in immunocompromised patients, but also related to nosocomial infections and even in healthy individuals. There is a widely variety of causal factors that contribute to yeast infection which means that candidiasis is a good example of a multifactorial syndrome.
- ↑ Tortora, Gerald, J. (2010). Mibrobiology:an Introduction. San Francisco, CA: Pearson Benjamin Cummings. p. 758.
- ↑ Mukherjee PK, Sendid B, Hoarau G, Colombel JF, Poulain D, Ghannoum MA (2015). "Mycobiota in gastrointestinal diseases". Nat Rev Gastroenterol Hepatol. 12 (2): 77–87. doi:10.1038/nrgastro.2014.188. PMID 25385227.
- ↑ Zadik Yehuda; Burnstein Saar; Derazne Estella; Sandler Vadim; Ianculovici Clariel; Halperin Tamar (March 2010). "Colonization of Candida: prevalence among tongue-pierced and non-pierced immunocompetent adults". Oral Dis. 16 (2): 172–5. doi:10.1111/j.1601-0825.2009.01618.x. PMID 19732353.
- ↑ Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 0-8385-8529-9.
- ↑ Tortora, Gerald, J. (2010). Microbiology: an Introduction. San Francisco, CA: Pearson Benjamin Cummings. p. 759.
- ↑ Ohama, T; Suzuki, Tsutomu; Mori, Miki; Osawa, Syozo; Ueda, Takuya; Watanabe, Kimitsuna; Nakase, Takashi (August 1993). "Non-universal decoding of the leucine codon CUG in several Candida species". Nucleic Acids Research. 21 (17): 1039–4045. doi:10.1093/nar/21.17.4039. PMC 309997
 . PMID 8371978.
. PMID 8371978. - ↑ Arnaud, MB; Costanzo, MC; Inglis, DO; Skrzypek, MS; Binkley, J; Shah, P; Binkley, G; Miyasato, SR; Sherlock, G. "CGD Help: Non-standard Genetic Codes". Candida Genome Database. Retrieved 30 October 2011.
- ↑ Andrzej (Anjay) Elzanowski and Jim Ostell (7 July 2010). "The Alternative Yeast Nuclear Code". The Genetic Codes. Bethesda, Maryland, U.S.A.: National Center for Biotechnology Information (NCBI). Retrieved 30 October 2011.
- ↑ Santos, MA; Cheesman, C; Costa, V; Moradas-Ferreira, P; Tuite, MF (February 1999). "Selective advantages created by codon ambiguity allowed for the evolution of an alternative genetic code in Candida spp.". Molecular Microbiology. 31 (3): 937–947. doi:10.1046/j.1365-2958.1999.01233.x. PMID 10048036.
- 1 2 Butler G, Rasmussen MD, Lin MF, et al. (June 2009). "Evolution of pathogenicity and sexual reproduction in eight Candida genomes". Nature. 459 (7247): 657–62. doi:10.1038/nature08064. PMC 2834264
 . PMID 19465905.
. PMID 19465905. - ↑ Silva RM, Paredes JA, Moura GR, et al. (October 2007). "Critical roles for a genetic code alteration in the evolution of the genus Candida". EMBO J. 26 (21): 4555–65. doi:10.1038/sj.emboj.7601876. PMC 2063480
 . PMID 17932489.
. PMID 17932489. - ↑ Peter E. Sudbery (2011). "Growth of Candida albicans hyphae" (PDF). Nature Reviews Microbiology. 9 (10): 737–748. doi:10.1038/nrmicro2636. PMID 21844880. See figure 2.
- ↑ Berman J, Sudbery PE (2002). "Candida Albicans: a molecular revolution built on lessons from budding yeast". Nature Reviews Genetics. 3 (12): 918–930. doi:10.1038/nrg948. PMID 12459722.
- ↑ Staib P, Morschhäuser J (2007). "Chlamydospore formation in Candida albicans and Candida dubliniensis--an enigmatic developmental programme.". Mycoses. 50 (1): 1–12. doi:10.1111/j.1439-0507.2006.01308.x. PMID 17302741.
- ↑ Rikkerrink E, Magee B, Magee P (1988). "Opaque-white phenotype transition: a programmed morphological transition in Candida albicans". J. Bact. 170 (2): 895–899. PMC 210739
 . PMID 2828333.
. PMID 2828333. - ↑ Soll D.R. (2012). Signal Transduction Pathways Regulating Switching, Mating and Biofilm Formation in Candida Albicans and Related Species. In: G. Witzany (ed). Biocommunication of Fungi. Springer, 85-102. ISBN 978-94-007-4263-5.
- ↑ Sonneborn A, Tebarth B, Ernst J (1999). "Control of white-opaque phenotypic switching in Candida albicans by the Efg1p morphogenetic regulator". Infection and Immunity. 67 (9): 4655–4660. PMC 96790
 . PMID 10456912.
. PMID 10456912. - ↑ Srikantha T, Tsai L, Daniels K, Soll D (2000). "EFG1 Null Mutants of Candida albicans Switch but Cannot Express the Complete Phenotype of White-Phase Budding Cells". J. Bact. 182 (6): 1580–1591. doi:10.1128/JB.182.6.1580-1591.2000. PMC 94455
 . PMID 10692363.
. PMID 10692363. - ↑ Larriba G, Calderone RA (2008). "Heterozygosity and Loss of Heterozygosity in Candida albicans". Pathogenic Fungi: Insights in Molecular Biology. Caister Academic Press. ISBN 978-1-904455-32-5.
- ↑ Staab, J. F. (1999). "Adhesive and Mammalian Transglutaminase Substrate Properties of Candida albicans Hwp1". Science. 283 (5407): 1535–1538. doi:10.1126/science.283.5407.1535. ISSN 0036-8075.
- ↑ Ariyachet, C.; Solis, N. V.; Liu, Y.; Prasadarao, N. V.; Filler, S. G.; McBride, A. E. (2013). "SR-like RNA-binding protein Slr1 affects Candida albicans filamentation and virulence". Infection and Immunity. 81 (4): 1267–1276. doi:10.1128/IAI.00864-12. ISSN 0019-9567.
- ↑ Duncan Wilson, Julian R. Naglik, and Bernhard Hube (2016). "The Missing Link between Candida albicans Hyphal Morphogenesis and Host Cell Damage". PLoS Pathog. PMC 5072684
 . PMID 27764260.
. PMID 27764260. - 1 2 Brosnahan, Mandy (July 22, 2013). "Candida Albicans". MicrobeWiki. Kenyon College.
- ↑ Sydnor, Emily (24 January 2011). "Hospital Epidemiology and Infection Control in Acute-Care Settings". Clinical Microbiology Reviews. 24: 141–173. doi:10.1128/CMR.00027-10. PMC 3021207
 . PMID 21233510.
. PMID 21233510. - ↑ Sardi, J. C. O. (2016-04-16). "Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options" (PDF). Journal of Medical Microbiology. doi:10.1099/jmm.0.045054-0. Retrieved 2016-04-16.
- ↑ Vazquez, Jose (2016-04-16). "Epidemiology, Management, and Prevention of Invasive Candidiasis". Medscape.org. Medscape. Retrieved 2016-04-16.
- ↑ Weinberger, M (2016-04-16). "Characteristics of candidaemia with Candida-albicans compared with non-albicans Candida species and predictors of mortality.". J Hosp Infect. doi:10.1016/j.jhin.2005.02.009. PMID 16009456.
- ↑ Yapar, Nur (2016-04-16). "Epidemiology and risk factors for invasive candidiasis". Dovepress. doi:10.2147/TCRM.S40160. PMC 3928396
 .
. - ↑ Di Giacomo, Raffaele; Maresca, Bruno; Porta, Amalia; Sabatino, Paolo; Carapella, Giovanni; Neitzert, Heinz-Christoph (2013). "Candida albicans/MWCNTs: A Stable Conductive Bio-Nanocomposite and Its Temperature-Sensing Properties". IEEE Transactions on Nanotechnology. 12 (2): 111. doi:10.1109/TNANO.2013.2239308.
- ↑ Rambach, G; Oberhauser, H; Speth, C; Lass-Flörl, C (2011). "Susceptibility of Candida species and various moulds to antimycotic drugs: Use of epidemiological cutoff values according to EUCAST and CLSI in an 8-year survey". Medical mycology : official publication of the International Society for Human and Animal Mycology. 49 (8): 856–63. doi:10.3109/13693786.2011.583943. PMID 21619497.
- ↑ Tortora. Microbiology an Introduction (tenth ed.). San Francisco,CA.: Pearson Benjamin Cummings. p. 759.
Further reading
- Waldman A, Gilhar A, Duek L, Berdicevsky I (May 2001). "Incidence of Candida in psoriasis—a study on the fungal flora of psoriatic patients". Mycoses. 44 (3–4): 77–81. doi:10.1046/j.1439-0507.2001.00608.x. PMID 11413927.
- Zordan RE, Miller MG, Galgoczy DJ, Tuch BB, Johnson AD (October 2007). "Interlocking Transcriptional Feedback Loops Control White-Opaque Switching in Candida albicans". PLoS Biology. 5 (10): e256. doi:10.1371/journal.pbio.0050256. PMC 1976629
 . PMID 17880264.
. PMID 17880264. - Rossignol T, Lechat P, Cuomo C, Zeng Q, Moszer I, d'Enfert C (January 2008). "CandidaDB: a multi-genome database for Candida species and related Saccharomycotina". Nucleic Acids Research. 36 (Database issue): D557–61. doi:10.1093/nar/gkm1010. PMC 2238939
 . PMID 18039716.
. PMID 18039716. - "How Candida albicans switches phenotype – and back again: the SIR2 silencing gene has a say in Candida's colony type". NCBI Coffeebreak. 1999-11-24. Retrieved 2008-11-02.
External links
| Wikimedia Commons has media related to Candida albicans. |
- Candida Genome Database
- U.S. National Institutes of Health on the Candida albicans genome
- Mycobank data on Candida albicans